A) The bond between carbon 1 of galactose and an oxygen atom of ceramide
B) The bond between carbon 1 of galactose and the oxygen atom in the galactose ring
C) The bond between carbon 4 of galactose and the oxygen atom in the galactose ring
D) The bond between carbon 4 of galactose and an oxygen atom of ceramide
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Fatty acid synthesis proceeds according to the following reaction: 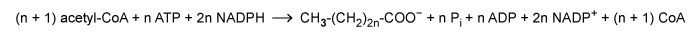 where n is a positive integer. Based on this, how many ADP and NADP+ molecules will be produced during synthesis of a 16-carbon fatty acid?
where n is a positive integer. Based on this, how many ADP and NADP+ molecules will be produced during synthesis of a 16-carbon fatty acid?
A) 7 ADP molecules and 14 NADP+ molecules
B) 14 ADP molecules and 28 NADP+ molecules
C) 8 ADP molecules and 15 NADP+ molecules
D) 16 ADP molecules and 32 NADP+ molecules
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
The cystic fibrosis transmembrane conductance regulator (CFTR) protein is a chloride ion channel involved in the production of mucus, sweat, and digestive fluids. CFTR is composed of five domains (Figure 1) : Two transmembrane domains (TMD1 and TMD2) , two nucleotide binding domains (NBD1 and NBD2) , and one regulatory domain (R domain) .
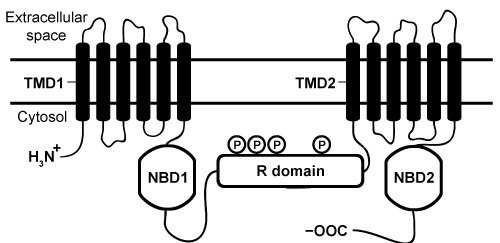 Figure 1 Schematic diagram of CFTR domainsThe NBD domains regulate CFTR activity by binding ATP, which causes a conformational change in the TMD domains. This change allows chloride ions to leave cells by passively crossing the membrane down their concentration gradient. The R domain regulates activity further through dynamic phosphorylation at several positions. The movement of chloride ions facilitates the movement of water out of the cell by osmotic pressure and helps keep secretions thin.Cystic fibrosis (CF) arises from recessively inherited mutations in the CFTR gene. The most common mutation is a three-base-pair deletion that removes a phenylalanine residue at position 508 in NBD1 (ΔF508) . This mutation causes the protein to fold incorrectly, making it more susceptible to degradation by proteases. The resulting decrease in CFTR abundance inhibits chloride ion transport, leading to thickening of normally thin secretions and increased risk of life-threatening pulmonary infections.
-Based on Figure 1, which of the following domains would most likely contain several isoleucine residues on its surface?
Figure 1 Schematic diagram of CFTR domainsThe NBD domains regulate CFTR activity by binding ATP, which causes a conformational change in the TMD domains. This change allows chloride ions to leave cells by passively crossing the membrane down their concentration gradient. The R domain regulates activity further through dynamic phosphorylation at several positions. The movement of chloride ions facilitates the movement of water out of the cell by osmotic pressure and helps keep secretions thin.Cystic fibrosis (CF) arises from recessively inherited mutations in the CFTR gene. The most common mutation is a three-base-pair deletion that removes a phenylalanine residue at position 508 in NBD1 (ΔF508) . This mutation causes the protein to fold incorrectly, making it more susceptible to degradation by proteases. The resulting decrease in CFTR abundance inhibits chloride ion transport, leading to thickening of normally thin secretions and increased risk of life-threatening pulmonary infections.
-Based on Figure 1, which of the following domains would most likely contain several isoleucine residues on its surface?
A) TMD1
B) NBD1
C) R domain
D) NBD2
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
The HIV-derived transactivator of transcription (Tat) is a short peptide with the sequence RKKRRQRRRPPQ. When paired with liposomes (ie, synthetic vesicles) composed of cationic lipids, Tat can transport large molecules across cell membranes and has been studied as a vector for gene therapy. However, Tat delivers cargo to cells with relatively low efficiency. Researchers hypothesized that a Tat homodimer could yield stronger DNA binding than the monomer, which may improve the efficiency of DNA delivery.To study this hypothesis, Tat and two variant peptides (Variants 1 and 2) were produced by solid-phase peptide synthesis, in which the growing peptide chain is immobilized on polystyrene beads. Unlike biological systems, solid-phase synthesis assembles peptides from the C-terminus to the N-terminus, as shown in Figure 1.
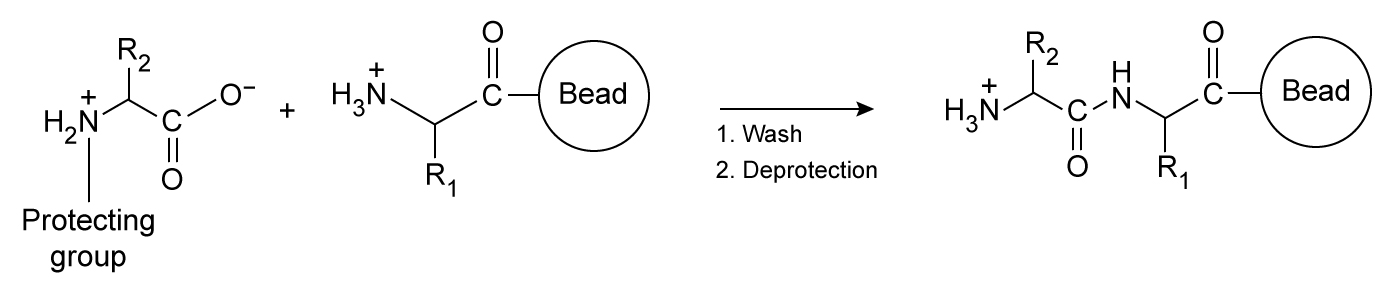 Figure 1 Simplified schematic of one round of solid-phase peptide synthesis. The protecting group is removed during deprotection. The process is repeated until the peptide is complete.Variants 1 and 2 contained a C-terminal and an N-terminal cysteine residue, respectively. Each variant was allowed to dimerize by disulfide bond formation. Peptide-DNA binding interactions were then studied by mixing varying amounts of each peptide with plasmid DNA encoding luciferase protein (pLuc) . The resulting peptide-DNA complexes were run on agarose gels, with results displayed in Figure 2.
Figure 1 Simplified schematic of one round of solid-phase peptide synthesis. The protecting group is removed during deprotection. The process is repeated until the peptide is complete.Variants 1 and 2 contained a C-terminal and an N-terminal cysteine residue, respectively. Each variant was allowed to dimerize by disulfide bond formation. Peptide-DNA binding interactions were then studied by mixing varying amounts of each peptide with plasmid DNA encoding luciferase protein (pLuc) . The resulting peptide-DNA complexes were run on agarose gels, with results displayed in Figure 2.
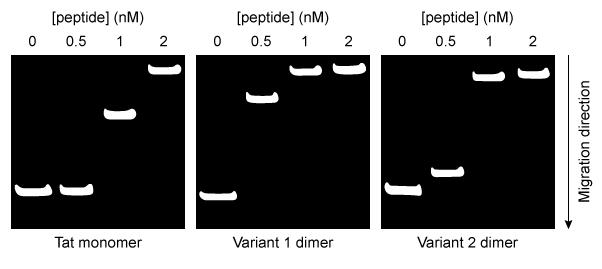 Figure 2 Agarose gel results for pLuc plasmid exposed to varying amounts of each peptideEqual concentrations of each type of peptide-DNA complex were then incubated with cationic liposomes and exposed to cultured cells for 24 hours. The cells were then assessed for luciferase activity. Greater luciferase activity indicated more efficient delivery of the pLuc plasmid. The results are shown in Figure 3.
Figure 2 Agarose gel results for pLuc plasmid exposed to varying amounts of each peptideEqual concentrations of each type of peptide-DNA complex were then incubated with cationic liposomes and exposed to cultured cells for 24 hours. The cells were then assessed for luciferase activity. Greater luciferase activity indicated more efficient delivery of the pLuc plasmid. The results are shown in Figure 3.
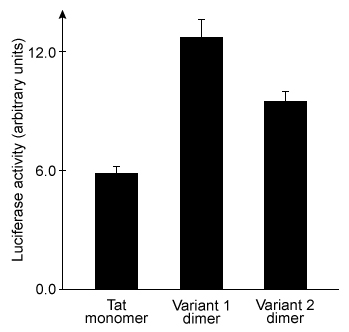 Figure 3 Plasmid delivery efficiency of three peptides, as measured by induced luciferase activity
Adapted from Lee S-J, Yoon S-H, Doh K-O. Enhancement of gene delivery using novel homodimeric tat peptide formed by disulfide bond. J Microbiol Biotechnol. 2011;21(8) :802-807.
-The addition of which reagent would result in the greatest amount of dimerization of the Tat variants?
Figure 3 Plasmid delivery efficiency of three peptides, as measured by induced luciferase activity
Adapted from Lee S-J, Yoon S-H, Doh K-O. Enhancement of gene delivery using novel homodimeric tat peptide formed by disulfide bond. J Microbiol Biotechnol. 2011;21(8) :802-807.
-The addition of which reagent would result in the greatest amount of dimerization of the Tat variants?
A) A strong acid such as HCl
B) A strong base such as NaOH
C) A reducing agent such as dithiothreitol
D) An oxidizing agent such as oxygen gas
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Upon entering muscle cells, glucose is immediately phosphorylated by hexokinase to produce glucose 6-phosphate (G6P) . This phosphorylation event prevents glucose from exiting the cell. G6P may then enter one of three metabolic pathways, depending on the needs of the cell (Figure 1) .
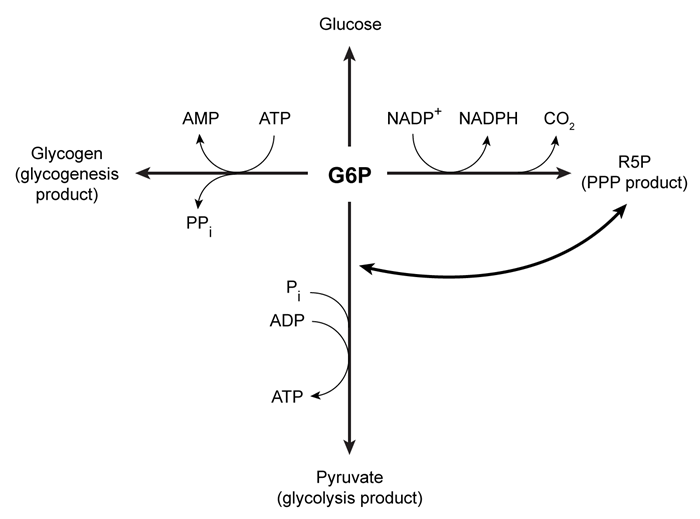 Figure 1 Three potential fates of G6P, determined by the needs of the cellUnder low ATP conditions, G6P enters glycolysis, where it is converted to two pyruvate molecules. The energy released in this process produces a net total of two ATP molecules per glucose molecule consumed. The resulting pyruvate molecules can then enter the mitochondria, where they participate in additional metabolic reactions that produce more ATP.When the cell experiences oxidative stress, G6P enters the pentose phosphate pathway (PPP) and produces NADPH, a reducing agent. During this process, G6P loses one carbon atom in the form of CO2 and is converted to ribose 5-phosphate (R5P) , which is required for nucleotide synthesis. In addition to low NADPH conditions, G6P may also enter the PPP during replication, when nucleotides are needed for DNA synthesis. If nucleotides are not needed but energy is, R5P can be converted back to glycolytic intermediates and reenter glycolysis. Each glucose molecule that is converted to R5P prior to glycolysis yields an average of 1.67 ATP molecules.When all the cell's metabolic requirements are met, additional G6P can enter glycogenesis and produce glycogen, which stores glucose for later use; this process consumes ATP. When signal molecules bind G protein-coupled receptors on muscle cells, glycogenolysis is activated to release individual monomers from glycogen, allowing them to enter the appropriate pathway.
-The glycogen branching enzyme forms a glycosidic bond between carbon 1 on the incoming glucose monomer and which of the following?
Figure 1 Three potential fates of G6P, determined by the needs of the cellUnder low ATP conditions, G6P enters glycolysis, where it is converted to two pyruvate molecules. The energy released in this process produces a net total of two ATP molecules per glucose molecule consumed. The resulting pyruvate molecules can then enter the mitochondria, where they participate in additional metabolic reactions that produce more ATP.When the cell experiences oxidative stress, G6P enters the pentose phosphate pathway (PPP) and produces NADPH, a reducing agent. During this process, G6P loses one carbon atom in the form of CO2 and is converted to ribose 5-phosphate (R5P) , which is required for nucleotide synthesis. In addition to low NADPH conditions, G6P may also enter the PPP during replication, when nucleotides are needed for DNA synthesis. If nucleotides are not needed but energy is, R5P can be converted back to glycolytic intermediates and reenter glycolysis. Each glucose molecule that is converted to R5P prior to glycolysis yields an average of 1.67 ATP molecules.When all the cell's metabolic requirements are met, additional G6P can enter glycogenesis and produce glycogen, which stores glucose for later use; this process consumes ATP. When signal molecules bind G protein-coupled receptors on muscle cells, glycogenolysis is activated to release individual monomers from glycogen, allowing them to enter the appropriate pathway.
-The glycogen branching enzyme forms a glycosidic bond between carbon 1 on the incoming glucose monomer and which of the following?
A) Carbon 3 on a glucose unit of the growing chain
B) Carbon 4 on a glucose unit of the growing chain
C) Carbon 5 on a glucose unit of the growing chain
D) Carbon 6 on a glucose unit of the growing chain
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Human gamma-glutamyl transpeptidase (hGGT) is an enzyme involved in cysteine homeostasis, and its overexpression has been linked to asthma, cancer, and other diseases. It catalyzes the addition of water across a bond to cleave extracellular glutathione (GSH) into glutamate and a dipeptide composed of cysteine and glycine. It is believed to function by the mechanism shown in Figure 1.
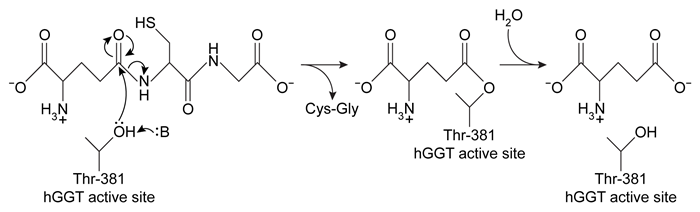 Figure 1 Proposed mechanism of hGGT-mediated GSH cleavageResearchers interested in investigating hGGT inhibitors as a treatment for pathologies related to excess hGGT activity performed the following experiments, each containing the same concentration of hGGT.Experiment 1hGGT was incubated with various concentrations of GSH in the presence or absence of 250-μM candidate compound acivicin or Compound 1. However, both candidates were found to be unsuitable as acivicin showed significant neurotoxicity and Compound 1 acted as an activator instead of an inhibitor.
Figure 1 Proposed mechanism of hGGT-mediated GSH cleavageResearchers interested in investigating hGGT inhibitors as a treatment for pathologies related to excess hGGT activity performed the following experiments, each containing the same concentration of hGGT.Experiment 1hGGT was incubated with various concentrations of GSH in the presence or absence of 250-μM candidate compound acivicin or Compound 1. However, both candidates were found to be unsuitable as acivicin showed significant neurotoxicity and Compound 1 acted as an activator instead of an inhibitor.
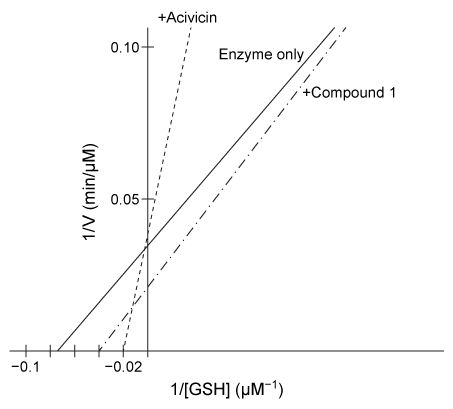 Figure 2 Lineweaver-Burk plot of hGGT activity with respect to GSH in the presence or absence of 250-μM candidate inhibitorsExperiment 2Researchers hypothesized that slight variations in the structure of Compound 1 could result in hGGT inhibition and tested several derivatives, shown in Table 1.Table 1 Kinetic Parameters of hGGT in the Presence or Absence of Several Candidate Inhibitors at 250 μM
Figure 2 Lineweaver-Burk plot of hGGT activity with respect to GSH in the presence or absence of 250-μM candidate inhibitorsExperiment 2Researchers hypothesized that slight variations in the structure of Compound 1 could result in hGGT inhibition and tested several derivatives, shown in Table 1.Table 1 Kinetic Parameters of hGGT in the Presence or Absence of Several Candidate Inhibitors at 250 μM
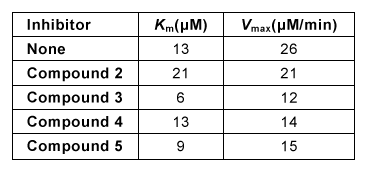 Experiment 3To better understand the physiological conditions under which hGGT operates, researchers exposed a cultured cell line expressing hGGT on the cell surface to a solution that mimicked the composition of extracellular fluid, including levels of GSH. They found that in this setup, uninhibited hGGT results in a reaction rate that is approximately ½Vmax.
Adapted from Wickham S, Regan N, West MB, et al. Inhibition of human ?-glutamyl transpeptidase: development of more potent, physiologically relevant, uncompetitive inhibitors. Biochem J. 2013;450(3) :547-57.
-In the reaction observed in Figure 1, hGGT acts as:
Experiment 3To better understand the physiological conditions under which hGGT operates, researchers exposed a cultured cell line expressing hGGT on the cell surface to a solution that mimicked the composition of extracellular fluid, including levels of GSH. They found that in this setup, uninhibited hGGT results in a reaction rate that is approximately ½Vmax.
Adapted from Wickham S, Regan N, West MB, et al. Inhibition of human ?-glutamyl transpeptidase: development of more potent, physiologically relevant, uncompetitive inhibitors. Biochem J. 2013;450(3) :547-57.
-In the reaction observed in Figure 1, hGGT acts as:
A) a hydrolase.
B) a transferase.
C) an isomerase.
D) a lyase.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Although the structure of the cell membrane has been investigated repeatedly, few studies examine the contribution of its lipid components. Lipid composition influences the fluidity and permeability of cell membranes in the human body. Chronic diseases such as atherosclerosis and psoriasis arise from alterations to the lipid composition.Atherosclerosis is caused by the accumulation of cholesterol in the endothelial cells lining the arterial wall. Cholesterol carried by low-density lipoprotein (LDL) is deposited into cell membranes, where it forms cholesterol crystalline domains (CCDs) . In damaged cells, these CCDs increase membrane permeability and contribute to the formation of cholesterol plaques that can potentially block the artery itself.Omega-3 fatty acids (O3FAs) derived from marine animals may be used to slow the development of atherosclerosis. Formulations of O3FA contain varying concentrations of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) (Figure 1) . Present research suggests that formulations of O3FA should contain EPA exclusively because it prevents cholesterol from organizing into CCDs. Nonetheless, atherosclerosis is generally treated by administering drugs referred to as statins, which decrease the free cholesterol in blood plasma by inhibiting the production of a cholesterol precursor. Statin therapy combined with O3FAs may be used to reduce atherosclerosis in patients with persistent hypertriglyceridemia (high triglyceride plasma levels) .
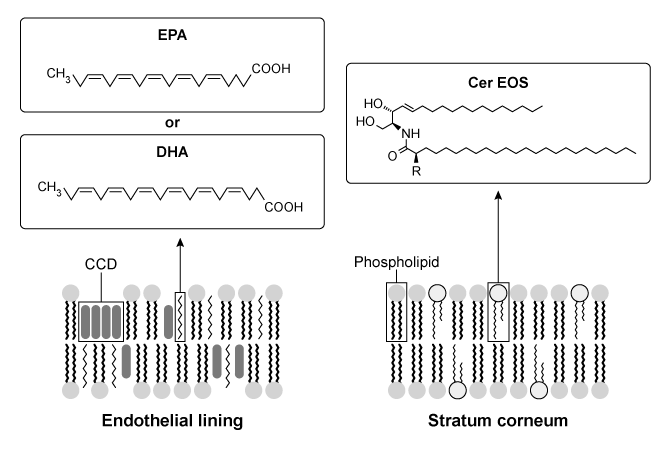 Figure 1 Organization of membranes in endothelial and epidermal cellsWater loss from the uppermost layer of the epidermis, the stratum corneum (SC) , is prevented by specialized lipids called ceramides (cer) . Psoriasis is a skin disease in which the permeability of the SC is compromised due to a significant decrease in the level of ceramides. The scarcity of ceramides impedes the formation of a lipid matrix called the long periodicity phase (LPP) in the cell membrane. Increasing the concentration of ceramides such as esterified omega-hydroxyacyl-sphingosine (cer-EOS) has been shown to reduce the permeability of SC cell membranes and may be used to treat psoriasis scabs (Figure 1) .
Adapted from Mason RP, Jacob RF, Shrivastava S, Sherratt SC, Chattopadhyay A. Eicosapentaenoic acid reduces membrane fluidity, inhibits cholesterol domain formation, and normalizes bilayer width in atherosclerotic-like model membranes. Biochim Biophys Acta. 2016;1858(12) :3131-3140.
-What structural feature in the molecules shown in Figure 1 would result in the greatest increase in cell membrane fluidity?
Figure 1 Organization of membranes in endothelial and epidermal cellsWater loss from the uppermost layer of the epidermis, the stratum corneum (SC) , is prevented by specialized lipids called ceramides (cer) . Psoriasis is a skin disease in which the permeability of the SC is compromised due to a significant decrease in the level of ceramides. The scarcity of ceramides impedes the formation of a lipid matrix called the long periodicity phase (LPP) in the cell membrane. Increasing the concentration of ceramides such as esterified omega-hydroxyacyl-sphingosine (cer-EOS) has been shown to reduce the permeability of SC cell membranes and may be used to treat psoriasis scabs (Figure 1) .
Adapted from Mason RP, Jacob RF, Shrivastava S, Sherratt SC, Chattopadhyay A. Eicosapentaenoic acid reduces membrane fluidity, inhibits cholesterol domain formation, and normalizes bilayer width in atherosclerotic-like model membranes. Biochim Biophys Acta. 2016;1858(12) :3131-3140.
-What structural feature in the molecules shown in Figure 1 would result in the greatest increase in cell membrane fluidity?
A) cis bond in EPA
B) trans bond in cer-EOS
C) Carboxyl group of DHA
D) Phosphate groups of phospholipids
Correct Answer

verified
Correct Answer
verified
Multiple Choice
The compound 5,10-methylene tetrahydrofolate (5,10mTHF) is derived from folic acid (vitamin B9) and is required in the active site of the enzyme thymidylate synthase for thymine synthesis. Therefore, 5,10mTHF is most likely:
A) a transcription factor.
B) an inhibitor.
C) an allosteric activator.
D) a coenzyme.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
The bacterium Clostridium difficile secretes protein toxins TcdA and TcdB, two homologues associated with C. difficile-related colitis. Tcd proteins enter host intestinal cells via endocytosis induced by interactions between the Tcd C-terminal domain and cell surface receptors. Endocytosis is followed by translocation of the N-terminal glucosyltransferase domain (GTD) across the endosomal membrane. The GTD remains anchored to endosomes by the central region of the protein, a transmembrane domain believed to form membrane pores and facilitate translocation. Inositol hexakisphosphate (IP6) binds Tcd proteins, an interaction that is stabilized by salt bridges in the binding pocket (Figure 1) .
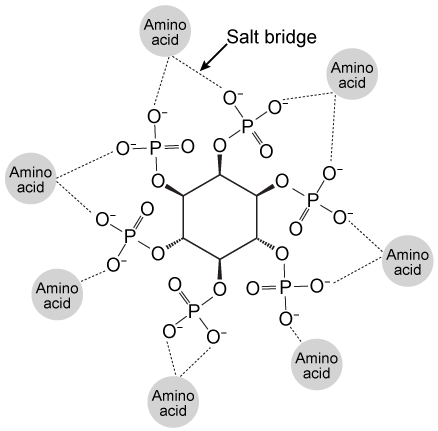 Figure 1 Salt bridges formed between IP6 and the amino acids in the binding pocketThis interaction induces a conformational change that causes a molecular flap to move away from the catalytic site. The catalytic site then mediates intramolecular cleavage in a reaction known as autoprocessing. This reaction results in the release of GTD into the cytosol, where it becomes active.To better understand the relationship between IP6 binding and catalytic activity, autoprocessing assays were performed. Researchers mutated histidine to alanine at positions 759 and 757 in the flap regions of TcdA and TcdB, respectively. They then compared the autoprocessing activities of H759A and H757A mutants to those of wild-type TcdA and TcdB at varying concentrations of IP6 (Figure 2) .
Figure 1 Salt bridges formed between IP6 and the amino acids in the binding pocketThis interaction induces a conformational change that causes a molecular flap to move away from the catalytic site. The catalytic site then mediates intramolecular cleavage in a reaction known as autoprocessing. This reaction results in the release of GTD into the cytosol, where it becomes active.To better understand the relationship between IP6 binding and catalytic activity, autoprocessing assays were performed. Researchers mutated histidine to alanine at positions 759 and 757 in the flap regions of TcdA and TcdB, respectively. They then compared the autoprocessing activities of H759A and H757A mutants to those of wild-type TcdA and TcdB at varying concentrations of IP6 (Figure 2) .
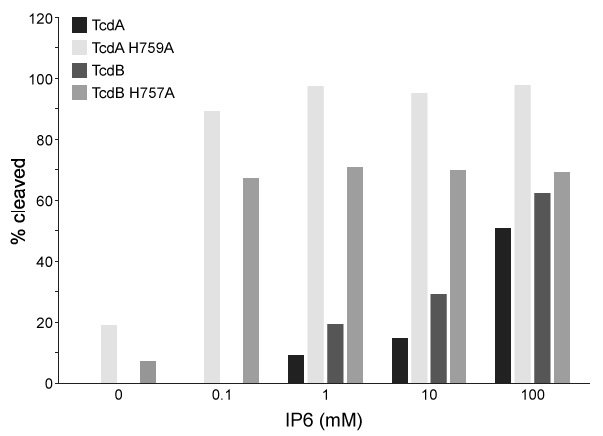 Figure 2 Percent cleavage of wild-type and mutant Tcd by autoprocessing
Adapted from Chumbler NM, Rutherford SA, Zhang Z, et al. Crystal structure of Clostridium difficile toxin A. Nat Microbiol. 2016;1:15002.
-Which of the following amino acid residues is LEAST likely to be present in the Tcd central region?
Figure 2 Percent cleavage of wild-type and mutant Tcd by autoprocessing
Adapted from Chumbler NM, Rutherford SA, Zhang Z, et al. Crystal structure of Clostridium difficile toxin A. Nat Microbiol. 2016;1:15002.
-Which of the following amino acid residues is LEAST likely to be present in the Tcd central region?
A) Ala
B) His
C) Val
D) Met
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Dystroglycan (Dg) is a transmembrane protein that mediates interactions between the cytoskeleton and the extracellular matrix (ECM) . It binds the ECM protein laminin in an interaction that is required for intercellular communication and muscle membrane structural stability. The Dg-laminin interaction is facilitated by a long carbohydrate chain called an O-mannosyl glycan, which is covalently linked to Dg (Figure 1) .
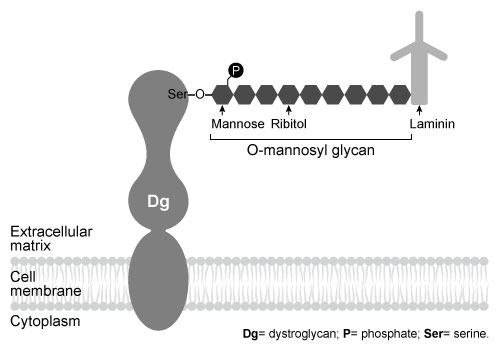 Figure 1 Schematic of interaction between Dg and laminin, mediated by the Dg-linked O-mannosyl glycanThe O-mannosyl glycan is added to Dg by a series of enzymes called glycosyltransferases. The first enzyme in this series, protein O-mannosyltransferase (POMT) , attaches D-mannose to the hydroxyl groups of serine and threonine residues in an α-linkage through the anomeric carbon. The next sugar in the chain then forms a β-1,4 linkage to the mannose residue, followed by several more steps to produce the full glycan. Genetic inactivation of any of the enzymes in the pathway results in an incomplete glycan and failure to bind laminin, and is the underlying cause of several severe forms of muscular dystrophy known as dystroglycanopathies.Researchers investigating dystroglycanopathies recently discovered the functions of two enzymes in the pathway: protein O-mannose kinase (POMK) and fukutin. POMK phosphorylates Dg-linked mannose at carbon 6, and fukutin adds the sugar ribitol 5-phosphate to the glycan chain. To assess the roles of POMK and fukutin relative to each other, scientists conducted experiments in which lysates from muscle cells deficient in either POMK or fukutin were incubated with 32P-labeled ATP or 3H-labeled activated ribitol 5-phosphate. Combined lysates containing both fukutin and POMK were also tested. Dg was then purified and detected by autoradiography and western blot using an antibody against Dg, as shown in Figure 2.
Figure 1 Schematic of interaction between Dg and laminin, mediated by the Dg-linked O-mannosyl glycanThe O-mannosyl glycan is added to Dg by a series of enzymes called glycosyltransferases. The first enzyme in this series, protein O-mannosyltransferase (POMT) , attaches D-mannose to the hydroxyl groups of serine and threonine residues in an α-linkage through the anomeric carbon. The next sugar in the chain then forms a β-1,4 linkage to the mannose residue, followed by several more steps to produce the full glycan. Genetic inactivation of any of the enzymes in the pathway results in an incomplete glycan and failure to bind laminin, and is the underlying cause of several severe forms of muscular dystrophy known as dystroglycanopathies.Researchers investigating dystroglycanopathies recently discovered the functions of two enzymes in the pathway: protein O-mannose kinase (POMK) and fukutin. POMK phosphorylates Dg-linked mannose at carbon 6, and fukutin adds the sugar ribitol 5-phosphate to the glycan chain. To assess the roles of POMK and fukutin relative to each other, scientists conducted experiments in which lysates from muscle cells deficient in either POMK or fukutin were incubated with 32P-labeled ATP or 3H-labeled activated ribitol 5-phosphate. Combined lysates containing both fukutin and POMK were also tested. Dg was then purified and detected by autoradiography and western blot using an antibody against Dg, as shown in Figure 2.
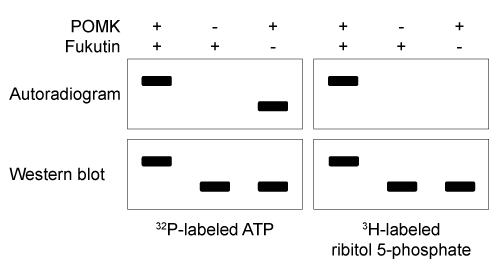 Figure 2 Autoradiograms and western blots of Dg purified from lysates incubated with 32P-labeled ATP or 3H-labeled ribitol 5-phosphate
Adapted from Kanagawa M, Kobayashi K, Tajiri M, et al. Identification of a Post-translational Modification with Ribitol-Phosphate and Its Defect in Muscular Dystrophy. Cell Rep. 2016;14(9) :2209-23.
-What was the purpose of the western blot in Figure 2?
Figure 2 Autoradiograms and western blots of Dg purified from lysates incubated with 32P-labeled ATP or 3H-labeled ribitol 5-phosphate
Adapted from Kanagawa M, Kobayashi K, Tajiri M, et al. Identification of a Post-translational Modification with Ribitol-Phosphate and Its Defect in Muscular Dystrophy. Cell Rep. 2016;14(9) :2209-23.
-What was the purpose of the western blot in Figure 2?
A) It confirmed the presence of Dg in all lanes (positive control) .
B) It showed that the molecular weight of Dg increases when enzymes are absent (dependent variable) .
C) It demonstrated experimental conditions for each lane (independent variable) .
D) It showed unmodified Dg (negative control) .
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Presynaptic nerve terminals release neurotransmitters via synaptic vesicle exocytosis. This action is mediated by the membrane-bound target-SNARE (t-SNARE) proteins syntaxin and SNAP-25, and the vesicle-associated SNARE (v-SNARE) protein synaptobrevin. Membrane fusion is initiated when the t-SNAREs and v-SNAREs form an α-helix bundle known as the core-trans-complex, achieved by zipping SNARE proteins together to form a more stable cis-complex (Figure 1) .
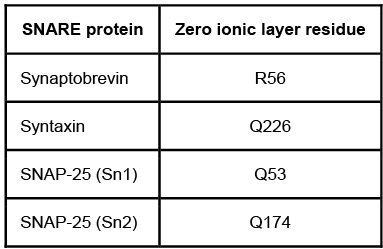
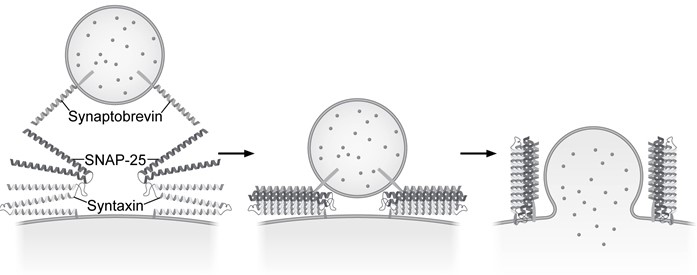 Figure 1 Formation of the core-trans-complex and subsequent membrane fusionThe cis-complex contains the zero ionic layer, the main site of interaction between complexed proteins. Within the zero ionic layer, arginine on synaptobrevin coordinates with carbonyl groups present on the other residues shown in Table 1. The zero ionic layer is buried within leucine zipper domains, which act as a shield to solvent molecules.Table 1 Composition of cis-Complex Zero Ionic Layer
Figure 1 Formation of the core-trans-complex and subsequent membrane fusionThe cis-complex contains the zero ionic layer, the main site of interaction between complexed proteins. Within the zero ionic layer, arginine on synaptobrevin coordinates with carbonyl groups present on the other residues shown in Table 1. The zero ionic layer is buried within leucine zipper domains, which act as a shield to solvent molecules.Table 1 Composition of cis-Complex Zero Ionic Layer
 An early step in cis-complex disassembly is destabilization by the ATPase N-ethylmaleimide-sensitive factor (NSF) , which initiates unzipping by breaking flanking leucine zipper regions, leading to vesicle reuptake. Movement of NSF to the cell membrane is mediated by the cytoplasmic protein α-SNAP, which must first bind the N-terminal domain of syntaxin.To determine the effect of vesicle-bound synaptobrevin on the binding of α-SNAP to syntaxin, increasing amounts of recombinant His-tagged α-SNAP were added to a constant amount of syntaxin affixed to glutathione-agarose beads with or without synaptobrevin. After incubation, bound proteins were recovered and quantitatively analyzed by immunoblotting. Results are shown in Figure 2.
An early step in cis-complex disassembly is destabilization by the ATPase N-ethylmaleimide-sensitive factor (NSF) , which initiates unzipping by breaking flanking leucine zipper regions, leading to vesicle reuptake. Movement of NSF to the cell membrane is mediated by the cytoplasmic protein α-SNAP, which must first bind the N-terminal domain of syntaxin.To determine the effect of vesicle-bound synaptobrevin on the binding of α-SNAP to syntaxin, increasing amounts of recombinant His-tagged α-SNAP were added to a constant amount of syntaxin affixed to glutathione-agarose beads with or without synaptobrevin. After incubation, bound proteins were recovered and quantitatively analyzed by immunoblotting. Results are shown in Figure 2.
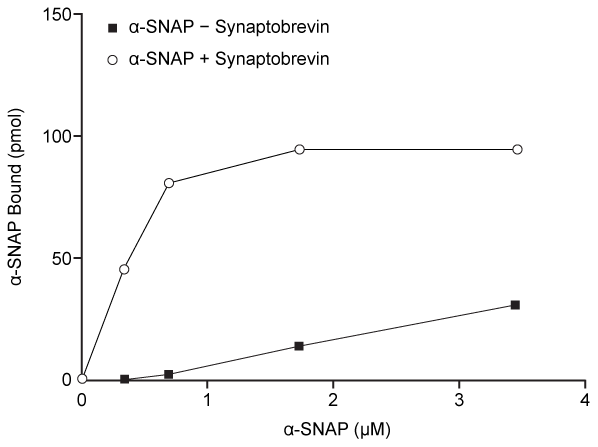 Figure 2 α-SNAP binding to syntaxin in the absence (square) or presence (circle) of synaptobrevin
Adapted from Mcmahon HT, Südhof TC. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity alpha-SNAP binding site. J Biol Chem. 1995;270(5) :2213-7.
-Which of the following statements regarding the binding of α-SNAP to syntaxin is best supported by the results shown in Figure 2?
Figure 2 α-SNAP binding to syntaxin in the absence (square) or presence (circle) of synaptobrevin
Adapted from Mcmahon HT, Südhof TC. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity alpha-SNAP binding site. J Biol Chem. 1995;270(5) :2213-7.
-Which of the following statements regarding the binding of α-SNAP to syntaxin is best supported by the results shown in Figure 2?
A) Binding does not occur in the presence of synaptobrevin.
B) Synaptobrevin induces a conformation change in syntaxin that results in lower binding affinity.
C) In the absence of synaptobrevin, binding does not saturate under the conditions of the experiment.
D) Interaction of synaptobrevin and syntaxin raises the Kd for the dissociation of syntaxin from α-SNAP.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Presynaptic nerve terminals release neurotransmitters via synaptic vesicle exocytosis. This action is mediated by the membrane-bound target-SNARE (t-SNARE) proteins syntaxin and SNAP-25, and the vesicle-associated SNARE (v-SNARE) protein synaptobrevin. Membrane fusion is initiated when the t-SNAREs and v-SNAREs form an α-helix bundle known as the core-trans-complex, achieved by zipping SNARE proteins together to form a more stable cis-complex (Figure 1) .
 Figure 1 Formation of the core-trans-complex and subsequent membrane fusionThe cis-complex contains the zero ionic layer, the main site of interaction between complexed proteins. Within the zero ionic layer, arginine on synaptobrevin coordinates with carbonyl groups present on the other residues shown in Table 1. The zero ionic layer is buried within leucine zipper domains, which act as a shield to solvent molecules.Table 1 Composition of cis-Complex Zero Ionic Layer
Figure 1 Formation of the core-trans-complex and subsequent membrane fusionThe cis-complex contains the zero ionic layer, the main site of interaction between complexed proteins. Within the zero ionic layer, arginine on synaptobrevin coordinates with carbonyl groups present on the other residues shown in Table 1. The zero ionic layer is buried within leucine zipper domains, which act as a shield to solvent molecules.Table 1 Composition of cis-Complex Zero Ionic Layer
 An early step in cis-complex disassembly is destabilization by the ATPase N-ethylmaleimide-sensitive factor (NSF) , which initiates unzipping by breaking flanking leucine zipper regions, leading to vesicle reuptake. Movement of NSF to the cell membrane is mediated by the cytoplasmic protein α-SNAP, which must first bind the N-terminal domain of syntaxin.To determine the effect of vesicle-bound synaptobrevin on the binding of α-SNAP to syntaxin, increasing amounts of recombinant His-tagged α-SNAP were added to a constant amount of syntaxin affixed to glutathione-agarose beads with or without synaptobrevin. After incubation, bound proteins were recovered and quantitatively analyzed by immunoblotting. Results are shown in Figure 2.
An early step in cis-complex disassembly is destabilization by the ATPase N-ethylmaleimide-sensitive factor (NSF) , which initiates unzipping by breaking flanking leucine zipper regions, leading to vesicle reuptake. Movement of NSF to the cell membrane is mediated by the cytoplasmic protein α-SNAP, which must first bind the N-terminal domain of syntaxin.To determine the effect of vesicle-bound synaptobrevin on the binding of α-SNAP to syntaxin, increasing amounts of recombinant His-tagged α-SNAP were added to a constant amount of syntaxin affixed to glutathione-agarose beads with or without synaptobrevin. After incubation, bound proteins were recovered and quantitatively analyzed by immunoblotting. Results are shown in Figure 2.
 Figure 2 α-SNAP binding to syntaxin in the absence (square) or presence (circle) of synaptobrevin
Adapted from Mcmahon HT, Südhof TC. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity alpha-SNAP binding site. J Biol Chem. 1995;270(5) :2213-7.
-During cis-complex disassembly, what consequence of NSF function is most likely responsible for destabilization of the cis-complex?
Figure 2 α-SNAP binding to syntaxin in the absence (square) or presence (circle) of synaptobrevin
Adapted from Mcmahon HT, Südhof TC. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity alpha-SNAP binding site. J Biol Chem. 1995;270(5) :2213-7.
-During cis-complex disassembly, what consequence of NSF function is most likely responsible for destabilization of the cis-complex?
A) Exposure of the zero ionic layer's charged amino acids to solvent molecules
B) Increased repulsive interactions among charged residues in the zero ionic layer
C) Increased amphipathic qualities of the leucine zipper dimer
D) Release of v-SNARE proteins from synaptic vesicles
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage The liver plays a central role in maintaining blood glucose homoeostasis. It contains the highest concentration of glycogen of any organ in the body and is one of few organs that can regenerate glucose from metabolic byproducts. The liver can replenish glucose in muscle and brain tissue during times of fasting, and it can help prevent lactate accumulation during intense physical activity.Von Gierke disease is a rare autosomal recessive disorder that arises from inactivating mutations in the liver enzyme glucose 6-phosphatase (G6Pase) . It affects roughly 1 in 100,000 individuals, and the resulting loss of both hormonal sensitivity and enzymatic activity leads to hypoglycemia (low blood sugar) . Von Gierke can also cause an increase in blood acidity concurrent with lactate build-up. Patients typically become symptomatic shortly after birth with convulsions, hyperventilation, and tremors. Left untreated, patients may develop gout, osteoporosis, and life-threatening complications such as kidney failure and liver tumors.In an effort to examine the biochemical consequences of this disorder, scientists generated a genetically engineered mouse model with liver G6Pase expression under the control of the Tet-Off system at both alleles. Under this system, addition of the small molecule tetracycline in the water source of the mouse rapidly shuts off G6Pase gene expression and therefore can recapitulate von Gierke disease. Blood glucose and pH levels were measured daily in two groups of genetically engineered mice over 20 days: group A mice received no tetracycline and group B mice were given tetracycline starting at day 10. All mice were fed identical low-sugar diets throughout the experiment. Adapted from Froissart R, Piraud M, Boudjemline AM, et al. Glucose-6-phosphatase deficiency. Orphanet J Rare Dis. 2011;6:27. -Which cofactor is regenerated by lactate synthesis?
A) NAD+
B) NADH
C) NADP+
D) NADPH
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate.
![Passage Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate. <strong>Figure 1</strong> AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.<strong>Table 1</strong> ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43. -What are the most likely role and effect of citrate on ACC activity? A) A cofactor that activates ACC B) A coenzyme that activates ACC C) An allosteric effector that activates ACC D) An inhibitor that inactivates ACC](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0dc2_af71_a3bf_719de41c80b7_MD0008_00.jpg) Figure 1 AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.Table 1 ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo
Figure 1 AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.Table 1 ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo
![Passage Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate. <strong>Figure 1</strong> AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.<strong>Table 1</strong> ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43. -What are the most likely role and effect of citrate on ACC activity? A) A cofactor that activates ACC B) A coenzyme that activates ACC C) An allosteric effector that activates ACC D) An inhibitor that inactivates ACC](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0dc2_af72_a3bf_e9a0f8dccdf8_MD0008_00.jpg) Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43.
-What are the most likely role and effect of citrate on ACC activity?
Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43.
-What are the most likely role and effect of citrate on ACC activity?
A) A cofactor that activates ACC
B) A coenzyme that activates ACC
C) An allosteric effector that activates ACC
D) An inhibitor that inactivates ACC
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Spinocerebellar ataxia 3 (SCA3) is a neurodegenerative disease that manifests as a progressive decrease in coordination of limbs and frequently results in impaired speech and eye coordination. It is associated with extension of the polyglutamine (polyQ) region of the ataxin-3 protein. The polyQ region is encoded by a series of consecutive CAG codons known as poly-CAG repeats. These regions vary in length from one person to another, ranging from as few as 10 to as many as 80 repeats. SCA3 has been reported in patients with polyQ regions exceeding ~55 repeats, and increased polyQ length correlates with earlier onset of the disease and greater clinical severity.Circular dichroism (CD) is a method of assessing protein secondary structure. It measures a molecule's ability to absorb left- and right-handed circularly polarized light of varying wavelengths. The absorption difference is known as "ellipticity," denoted by Δε. Alpha-helices have a maximum Δε at 190 nm, and beta-sheets have a maximum Δε at 200 nm.CD results show that as the length of the ataxin-3 polyQ region increases, alpha-helical nature is lost and the protein aggregates, ultimately precipitating out of solution and depositing in brain and other tissues. Furthermore, as proteins aggregate, they form beta-sheets. These beta-sheets are in the parallel orientation, and each sheet can interact with sheets in other proteins to form long, insoluble structures known as amyloid fibers (Figure 1) .
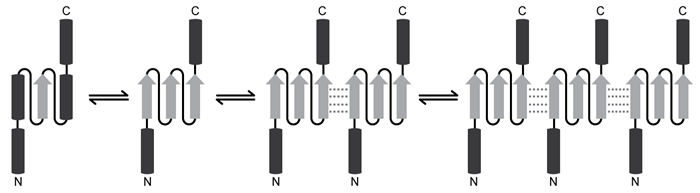 Figure 1 Depiction of the ataxin-3 polyQ region shifting from alpha-helical (cylinders) to beta-sheet (arrows) structure and aggregatingGlutamine can be encoded by both CAG and CAA codons. Researchers investigated survival rates in fruit flies expressing ataxin-3 with polyQ regions of equal length encoded by either CAG-CAG or CAA-CAG repeats (Table 1) . The protein products had identical amino acid sequences.Table 1 Fruit Fly Survival Rates Relative to Ataxin-3 Expression
Figure 1 Depiction of the ataxin-3 polyQ region shifting from alpha-helical (cylinders) to beta-sheet (arrows) structure and aggregatingGlutamine can be encoded by both CAG and CAA codons. Researchers investigated survival rates in fruit flies expressing ataxin-3 with polyQ regions of equal length encoded by either CAG-CAG or CAA-CAG repeats (Table 1) . The protein products had identical amino acid sequences.Table 1 Fruit Fly Survival Rates Relative to Ataxin-3 Expression
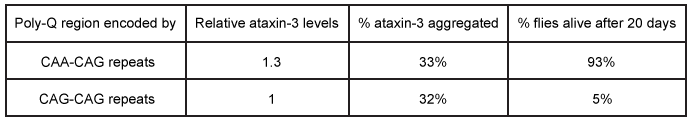 Adapted from Bevivino AE, Loll PJ. An expanded glutamine repeat destabilizes native ataxin-3 structure and mediates formation of parallel beta -fibrils. Proc Natl Acad Sci USA. 2001;98(21) :11955-60.
-Why might ataxin-3 aggregates become insoluble?
Adapted from Bevivino AE, Loll PJ. An expanded glutamine repeat destabilizes native ataxin-3 structure and mediates formation of parallel beta -fibrils. Proc Natl Acad Sci USA. 2001;98(21) :11955-60.
-Why might ataxin-3 aggregates become insoluble?
A) The change in conformation from alpha-helix to beta-sheet exposes hydrophobic residues.
B) The conformational change from alpha-helix to beta-sheet disrupts side-chain ionic interactions.
C) Interactions between glutamine side chains exclude them from interacting with water.
D) Beta-sheets expose more of the hydrophobic protein backbone than do alpha-helices.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
The Ankyrin repeat and SOCS box containing protein 4 (Asb-4) is highly expressed in the cytosol of hypothalamus cells during the well-fed state but is downregulated during fasting. In addition, the protein insulin receptor substrate 4 (IRS4) , known to be involved in appetite regulation, is expressed in the cytosol of the same cells as Asb-4. Accordingly, it was hypothesized that Asb-4 suppresses appetite through interactions with IRS4.Small peptides such as myc (EQKLISEEDL) and flag (DYKDDDDK) are often attached to recombinant proteins to facilitate their purification and detection. HEK293 cells were transfected with plasmids encoding Asb-4 and IRS4 tagged with myc or flag at their N-termini (myc-Asb4 and flag-IRS4, respectively) . Cell lysates were incubated with beads linked to α-flag, an antibody that binds flag, and proteins that did not bind were washed away. The bound proteins were then visualized by Western blot using α-myc antibodies. The α-myc antibody detected the presence of myc, indicating that myc-Asb-4 binds to flag-IRS4.To better understand interactions between Asb-4 and IRS4, cells were transfected with 1 μg of flag-IRS4 plasmid and varying amounts of either myc-Asb-4 plasmid or a plasmid with the sb domain deleted (myc-Asb-4Δsb) . Flag-IRS4 levels from each set of cells were then quantified, as shown in Figure 1.
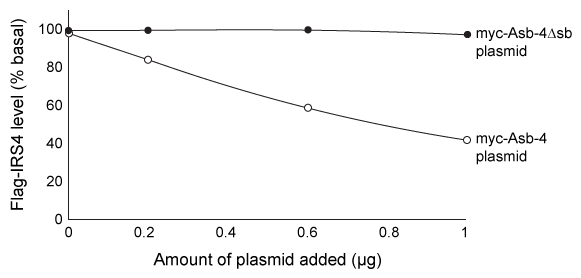 Figure 1 Flag-IRS4 levels in cells with varying amounts of myc-Asb-4 or myc-Asb-4Δsb plasmid addedIn a follow-up experiment, lysates from transfected cells were again incubated with the α-flag beads, and a Western blot was performed on the precipitated proteins using α-flag, α-myc, and an antibody against ubiquitin (α-ub) . The results are shown in Figure 2.
Figure 1 Flag-IRS4 levels in cells with varying amounts of myc-Asb-4 or myc-Asb-4Δsb plasmid addedIn a follow-up experiment, lysates from transfected cells were again incubated with the α-flag beads, and a Western blot was performed on the precipitated proteins using α-flag, α-myc, and an antibody against ubiquitin (α-ub) . The results are shown in Figure 2.
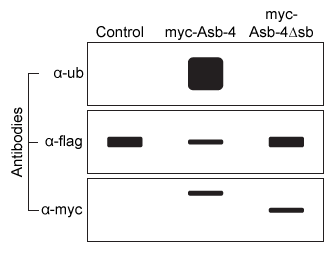 Figure 2 Ubiquitin, flag-IRS4, and myc-Asb-4 levels in cells transfected with flag-IRS4 and either an empty vector (control) , myc-Asb-4 plasmid, or myc-Asb-4Δsb plasmid
Adapted from Li JY, Chai B, Zhang W, et al. Ankyrin repeat and SOCS box containing protein 4 (Asb-4) colocalizes with insulin receptor substrate 4 (IRS4) in the hypothalamic neurons and mediates IRS4 degradation. BMC Neurosci. 2011;12:95.
-Which of the following most accurately describes the interaction between α-flag and flag-IRS4?
Figure 2 Ubiquitin, flag-IRS4, and myc-Asb-4 levels in cells transfected with flag-IRS4 and either an empty vector (control) , myc-Asb-4 plasmid, or myc-Asb-4Δsb plasmid
Adapted from Li JY, Chai B, Zhang W, et al. Ankyrin repeat and SOCS box containing protein 4 (Asb-4) colocalizes with insulin receptor substrate 4 (IRS4) in the hypothalamic neurons and mediates IRS4 degradation. BMC Neurosci. 2011;12:95.
-Which of the following most accurately describes the interaction between α-flag and flag-IRS4?
A) A noncovalent protein-protein interaction
B) A covalent protein-protein interaction
C) A noncovalent protein-carbohydrate interaction
D) A covalent protein-carbohydrate interaction
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
The influenza A virus is an enveloped RNA virus with potential for zoonosis (transmission from animals to humans) . It obtains its plasma membrane from its host cell, but the composition of the viral envelope and vertebrate plasma membrane are distinct. The envelope of H6-G225D consists mostly of phosphatidylethanolamine as well as some negatively charged glycerophospholipids. The outer membrane contains hemagglutinin (HA) proteins that bind to the carbohydrate known as sialic acid (SA) on host cell receptor proteins, facilitating viral entry. It also contains neuraminidase enzymes that break the bond between SA and galactose in the host membrane glycans, allowing new virions to be released.Minute changes in the genes that code for HA proteins can alter the virulence, host, and attachment mechanism. In general, influenza A infects avian species by attaching to tracheal tissue, which displays SA groups bound to receptors through an α-2,3-glycosidic linkage to galactose (Figure 1) . Avian viruses generally cannot attack human cells, in which SA is linked to galactose through an α-2,6-glycosidic linkage instead of an α-2,3 linkage.
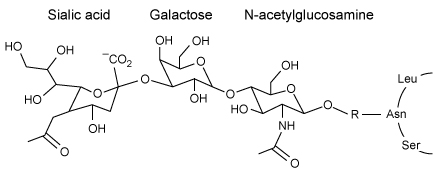 Figure 1 Structure of the influenza A receptor in avian cell membranesIn a recent study, however, a G225D substitution in HA, caused by a GGC to GAU codon mutation, enabled avian virions to infect human cells. The stability of G225D HA proteins was compared to that of H6-P186L, which is created by a single CCC to CUC mutation and exclusively infects avian cells, and H1N1, which targets only humans. The viral RNA of H6-P186L, H6-G225D, and H1N1 was cloned using cDNA and expressed in embryonic kidney cells. The melting curve of each purified HA protein was determined using differential scanning calorimetry, with peaks representing melting temperatures (Figure 2) .
Figure 1 Structure of the influenza A receptor in avian cell membranesIn a recent study, however, a G225D substitution in HA, caused by a GGC to GAU codon mutation, enabled avian virions to infect human cells. The stability of G225D HA proteins was compared to that of H6-P186L, which is created by a single CCC to CUC mutation and exclusively infects avian cells, and H1N1, which targets only humans. The viral RNA of H6-P186L, H6-G225D, and H1N1 was cloned using cDNA and expressed in embryonic kidney cells. The melting curve of each purified HA protein was determined using differential scanning calorimetry, with peaks representing melting temperatures (Figure 2) .
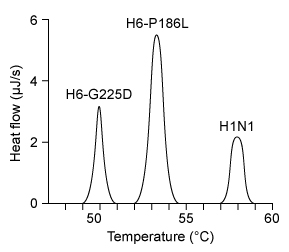 Figure 2 Hemagglutinin melting curves for H6N1 and H1N1 influenza A strains
Adapted from De vries RP, Tzarum N, Peng W, et al. A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors. EMBO Mol Med. 2017;9(9) :1314-1325.
-The absence of which of the following lipids is most likely to affect the assembly of viral envelopes?
Figure 2 Hemagglutinin melting curves for H6N1 and H1N1 influenza A strains
Adapted from De vries RP, Tzarum N, Peng W, et al. A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors. EMBO Mol Med. 2017;9(9) :1314-1325.
-The absence of which of the following lipids is most likely to affect the assembly of viral envelopes?
A) Palmitic acid
B) Triacylglycerides
C) Sphingomyelin
D) Phosphatidylserine
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Physical activity produces an increased energy requirement in the muscles as well as in the brain. During periods of intense exercise, glycogenolysis is activated in muscles to provide them with the energy needed for contractions, and research indicates that astrocytes also consume glycogen at increased levels during exercise. It is hypothesized that during periods of physical exertion, astrocytes convert glycogen to lactate, which is then used as a fuel source in neurons.Monocarboxylate transporters (MCTs) are responsible for the movement of lactate across membranes, and MCT2 is expressed at increased levels in neuronal cells during exercise. Scientists proposed that lactate is secreted from astrocytes and taken up by neurons through MCT2. Lactate is then converted to pyruvate and enters the mitochondria, where it is further metabolized. To test their predictions, scientists examined the effect of exercise on glycogen metabolism in rodents.Experiment 1Rats were either allowed to rest or exercised to exhaustion. The levels of several metabolites were then quantified in both the plantaris muscle and the hippocampus.
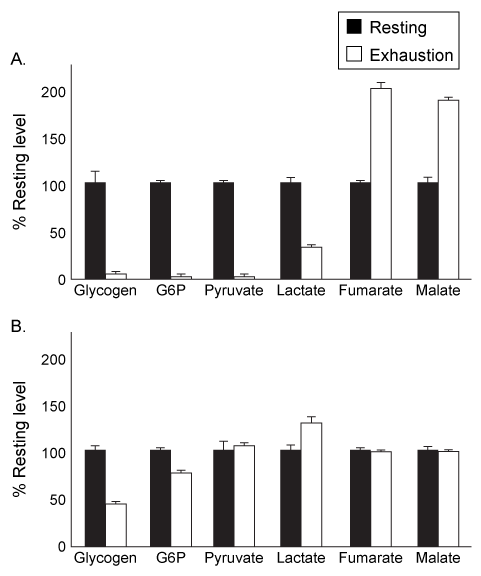 Figure 1 Levels of several metabolites in plantaris muscle (A) and hippocampus (B) from rats at rest or after exercising to exhaustionExperiment 2Rats were injected with 1,4-dideoxy-1,4-imino-D-arabinitol (DAB) or saline, and glycogen and lactate levels were measured in the hippocampus of rats at rest or after exercise to exhaustion.
Figure 1 Levels of several metabolites in plantaris muscle (A) and hippocampus (B) from rats at rest or after exercising to exhaustionExperiment 2Rats were injected with 1,4-dideoxy-1,4-imino-D-arabinitol (DAB) or saline, and glycogen and lactate levels were measured in the hippocampus of rats at rest or after exercise to exhaustion.
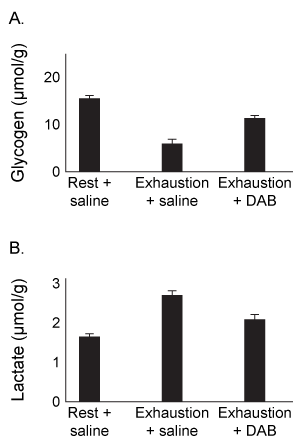 Figure 2 Levels of glycogen (A) and lactate (B) in rats injected with DAB or saline, after rest or upon exhaustion
Adapted from Matsui T, Omuro H, Liu Y-F, et al. Astrocytic glycogen-derived lactate fuels the brain during exhaustive exercise to maintain endurance capacity. Proceedings of the National Academy of Sciences. 2017;114(24) :6358-6363.
-The intermediates of which metabolic pathway were shown to increase in muscles due to exercise?
Figure 2 Levels of glycogen (A) and lactate (B) in rats injected with DAB or saline, after rest or upon exhaustion
Adapted from Matsui T, Omuro H, Liu Y-F, et al. Astrocytic glycogen-derived lactate fuels the brain during exhaustive exercise to maintain endurance capacity. Proceedings of the National Academy of Sciences. 2017;114(24) :6358-6363.
-The intermediates of which metabolic pathway were shown to increase in muscles due to exercise?
A) The citric acid cycle
B) Fermentation
C) Glycolysis
D) Glycogenolysis
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Which experimental procedure(s) must scientists use to determine Vmax and Km of an enzymatic reaction using the Michaelis-Menten model? They must ensure that:they only measure the initial reaction rate for each substrate concentration.total enzyme concentration is much greater than the Km of the reaction.each initial substrate concentration tested is much greater than enzyme concentration.
A) I only
B) III only
C) I and II only
D) I and III only
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
The tumor suppressor p53 is a homotetrameric protein with an N-terminal transactivation domain (TAD) , a proline-rich domain, a DNA-binding domain (DBD) , a tetramerization domain (TD) , and a C-terminal regulatory domain (REG) on each subunit (Figure 1) .
 Figure 1 Organization of the p53 domains (1-393 amino acids) Human double minute 2 (Hdm2) is a lysine-specific ubiquitin protein ligase that can change the half-life of some proteins, including p53, by signaling for their proteolytic cleavage. Under normal circumstances, Hdm2 binds p53 at the AD1 region within the TAD and attaches ubiquitin to a lysine residue. However, errors in DNA replication induce the phosphorylation of residue T18 on p53, which inhibits p53's association with Hdm2. The overexpression of Hdm2 has been correlated with the downregulation of tumor suppressor p53 in certain types of cancers. Changes in the NMR chemical shift (ppm) of amino acids in the AD1 region shown in Figure 2 provide evidence for the formation of a p53-Hdm2 complex.
Figure 1 Organization of the p53 domains (1-393 amino acids) Human double minute 2 (Hdm2) is a lysine-specific ubiquitin protein ligase that can change the half-life of some proteins, including p53, by signaling for their proteolytic cleavage. Under normal circumstances, Hdm2 binds p53 at the AD1 region within the TAD and attaches ubiquitin to a lysine residue. However, errors in DNA replication induce the phosphorylation of residue T18 on p53, which inhibits p53's association with Hdm2. The overexpression of Hdm2 has been correlated with the downregulation of tumor suppressor p53 in certain types of cancers. Changes in the NMR chemical shift (ppm) of amino acids in the AD1 region shown in Figure 2 provide evidence for the formation of a p53-Hdm2 complex.
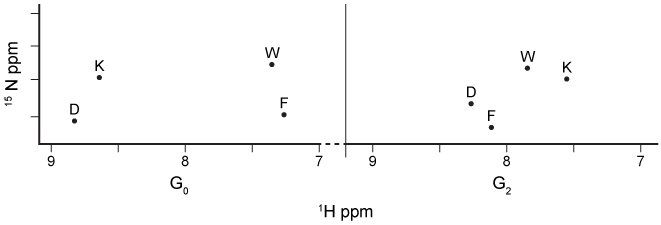 Figure 2 1H NMR spectrum of the AD1 region in 15N-labeled p53 during G0 and G2 phases of the cell cycleHdm2 also binds to Nedd8 ultimate buster 1 (NUB1) , which enhances signaling for the degradation of Nedd8 proteins. Preliminary data suggest that Hdm2 interacts differently with wild-type NUB1 (WTNUB1) than with mutant K159RNUB1. To assess the effect of Hdm2 on NUB1 and p53, the half-lives of p53 and Nedd8 protein variants were compared in cells that overexpress Hdm2 (Figure 3) .
Figure 2 1H NMR spectrum of the AD1 region in 15N-labeled p53 during G0 and G2 phases of the cell cycleHdm2 also binds to Nedd8 ultimate buster 1 (NUB1) , which enhances signaling for the degradation of Nedd8 proteins. Preliminary data suggest that Hdm2 interacts differently with wild-type NUB1 (WTNUB1) than with mutant K159RNUB1. To assess the effect of Hdm2 on NUB1 and p53, the half-lives of p53 and Nedd8 protein variants were compared in cells that overexpress Hdm2 (Figure 3) .
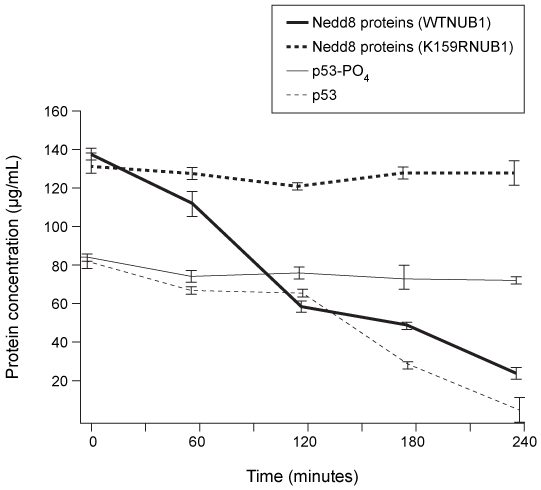 Figure 3 Half-lives of p53, phosphorylated p53 (p53-PO4) , and Nedd-8 proteins in cells with wild-type or mutant NUB1 over time
Adapted from Ferreon JC, Lee CW, Arai M, Martinez-yamout MA, Dyson HJ, Wright PE. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci USA. 2009;106(16) :6591-6.
-P53 displays positive cooperativity when it binds to promoters on DNA. Which statement regarding p53 structure and function accurately explains this observation?
Figure 3 Half-lives of p53, phosphorylated p53 (p53-PO4) , and Nedd-8 proteins in cells with wild-type or mutant NUB1 over time
Adapted from Ferreon JC, Lee CW, Arai M, Martinez-yamout MA, Dyson HJ, Wright PE. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci USA. 2009;106(16) :6591-6.
-P53 displays positive cooperativity when it binds to promoters on DNA. Which statement regarding p53 structure and function accurately explains this observation?
A) The DBD subunits bind individually to DNA and have independent binding affinities.
B) Phosphorylation of the REG increases the affinity of the DBD for DNA.
C) The affinity of one DBD subunit for DNA increases as adjacent subunits bind to the DNA.
D) Once the first promoter is bound, the binding affinity of the DBD decreases.
Correct Answer

verified
Correct Answer
verified
Showing 161 - 180 of 307