A) GL2
B) GL3
C) GL5
D) GL6
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
NDPK is an enzyme in budding yeast that uses ATP to phosphorylate nucleotide diphosphates (NDPs and dNDPs) , yielding the triphosphate form of each (NTPs and dNTPs) . dNTPs are necessary for DNA synthesis, yet yeast cells that lack NDPK grow and divide at the same rate as wild-type yeast. Therefore, triphosphate generation is most likely carried out by one or more other enzymes.The enzyme TMPK converts deoxythymidine monophosphate (dTMP) to the diphosphate form (dTDP) by transferring the γ-phosphate of ATP to dTMP. Yeast cells that lack TMPK activity cannot synthesize DNA and become arrested during replication. It was proposed that TMPK may partially compensate for NDPK loss by phosphorylating dTDP as well as dTMP.To better understand the role of TMPK in nucleotide phosphorylation, wild-type TMPK and the inactive E75K variant were purified by affinity chromatography, and the following experiments were performed.Experiment 1After purification, a sample containing 3 μg of protein was added to a reaction mixture containing radiolabeled nucleotide monophosphates (NMPs or dNMPs) and unlabeled ATP. The reaction proceeded for 30 minutes, and products were separated by thin-layer chromatography (TLC) on a PEI-cellulose plate. PEI-cellulose separates nucleotides by charge (Figure 1) , and migration distance decreases with increasing negative charge. Spots were visualized by autoradiography, and it was determined that 0.30 nmol of dTMP and 0.15 nmol of uridine monophosphate (UMP) became phosphorylated, respectively.
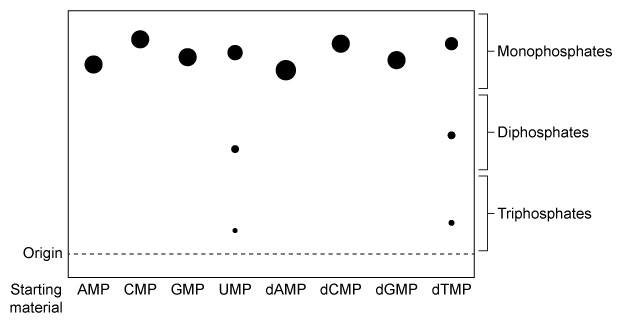 Figure 1 TLC analysis of nucleotide monophosphates after exposure to TMPK and ATP. (Note: Different nucleotides with the same phosphorylation level migrate different distances.) Experiment 2The experiment was repeated using radiolabeled dTDP as the starting material. The reaction proceeded in buffer only, in the presence of E75K TMPK, or in the presence of wild-type TMPK. Products were separated by TLC and visualized (Figure 2) .
Figure 1 TLC analysis of nucleotide monophosphates after exposure to TMPK and ATP. (Note: Different nucleotides with the same phosphorylation level migrate different distances.) Experiment 2The experiment was repeated using radiolabeled dTDP as the starting material. The reaction proceeded in buffer only, in the presence of E75K TMPK, or in the presence of wild-type TMPK. Products were separated by TLC and visualized (Figure 2) .
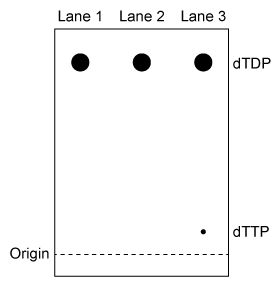 Figure 2 TLC analysis of dTDP phosphorylation with buffer only (Lane 1) , E75K TMPK (Lane 2) , or wild-type TMPK (Lane 3)
Adapted from Chien CY, Chen BR, Chou CK, Sclafani RA, Su JY. The yeast Cdc8 exhibits both deoxythymidine monophosphate and diphosphate kinase activities. FEBS Lett. 2009;583(13) :2281-6.
-A Mg2+ ion in the active site of TMPK is directly involved in catalysis. Given this, Mg2+ most likely plays which of the following roles in TMPK-mediated reactions?
Figure 2 TLC analysis of dTDP phosphorylation with buffer only (Lane 1) , E75K TMPK (Lane 2) , or wild-type TMPK (Lane 3)
Adapted from Chien CY, Chen BR, Chou CK, Sclafani RA, Su JY. The yeast Cdc8 exhibits both deoxythymidine monophosphate and diphosphate kinase activities. FEBS Lett. 2009;583(13) :2281-6.
-A Mg2+ ion in the active site of TMPK is directly involved in catalysis. Given this, Mg2+ most likely plays which of the following roles in TMPK-mediated reactions?
A) It binds negatively charged amino acids, stabilizing the tertiary structure of TMPK.
B) It interacts with water molecules in the active site, facilitating phosphate hydrolysis.
C) It binds anions, preventing them from competing with phosphate groups for the active site.
D) It interacts with the negative charges of phosphate groups, stabilizing the transition state.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Many cephalopods can change color to blend in with their surroundings by altering the conformations of particular proteins within their skin cells. The reflectin proteins play a major role in this camouflage mechanism. Reflectins are a group of intrinsically disordered proteins that do not adopt clearly defined secondary or tertiary structures. Under certain conditions reflectins aggregate, allowing cells to reflect various wavelengths of light.Skin cells from the squid Doryteuthis opalescens were cultured and three reflectins-DoRA, DoRB, and DoRC-were purified and analyzed to determine amino acid composition. They were found to contain an abundance of arginine and serine but relatively few cysteine, leucine, isoleucine, valine, phenylalanine, or tryptophan residues. Figure 1 shows the relative hydrophobicity of each reflectin variant as a function of amino acid position.
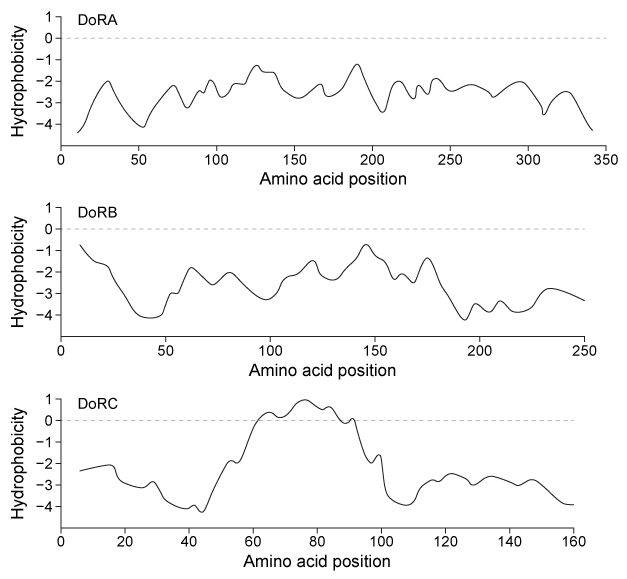 Figure 1 Hydrophobicity plots of three reflectins isolated from D. opalescens. (Note: Negative numbers indicate hydrophilic regions.) In D. opalescens skin cells (pH ≈ 7.8) , reflectins aggregate in response to acetylcholine (ACh) , which causes a signal cascade that changes the phosphorylation state of the proteins. Cells were incubated with or without ACh, and DoRA and DoRB were purified and assessed for phosphorylation by mass spectrometry. DoRA became phosphorylated in the presence of ACh whereas DoRB became dephosphorylated. After examining the isoelectric points of unmodified DoRA and DoRB, as shown in Figure 2, scientists proposed that reflectins aggregate upon charge neutralization.
Figure 1 Hydrophobicity plots of three reflectins isolated from D. opalescens. (Note: Negative numbers indicate hydrophilic regions.) In D. opalescens skin cells (pH ≈ 7.8) , reflectins aggregate in response to acetylcholine (ACh) , which causes a signal cascade that changes the phosphorylation state of the proteins. Cells were incubated with or without ACh, and DoRA and DoRB were purified and assessed for phosphorylation by mass spectrometry. DoRA became phosphorylated in the presence of ACh whereas DoRB became dephosphorylated. After examining the isoelectric points of unmodified DoRA and DoRB, as shown in Figure 2, scientists proposed that reflectins aggregate upon charge neutralization.
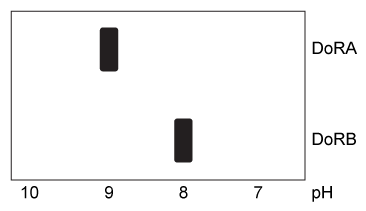 Figure 2 Isoelectric focusing of DoRA and DoRB
Adapted from Demartini DG, Izumi M, Weaver AT, Pandolfi E, Morse DE. Structures, Organization, and Function of Reflectin Proteins in Dynamically Tunable Reflective Cells. J Biol Chem. 2015;290(24) :15238-49.
-Which reason best explains why reflectin proteins do not adopt a defined tertiary structure?
Figure 2 Isoelectric focusing of DoRA and DoRB
Adapted from Demartini DG, Izumi M, Weaver AT, Pandolfi E, Morse DE. Structures, Organization, and Function of Reflectin Proteins in Dynamically Tunable Reflective Cells. J Biol Chem. 2015;290(24) :15238-49.
-Which reason best explains why reflectin proteins do not adopt a defined tertiary structure?
A) Reflectins are unable to interact with water efficiently.
B) Reflectins only experience a weak hydrophobic effect.
C) Reflectins cannot participate in hydrogen bonding.
D) Reflectins are highly basic and therefore cannot fold.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Aldehyde dehydrogenase 2 (ALDH2) is essential for the oxidation of toxic aldehydes, and uses NAD+ as the oxidizing agent. Mitochondrial ALDH2 exists as a 220-kD homotetrameric protein containing a catalytic, nucleotide-binding, and coenzyme-binding domain. The most prevalent ALDH2 mutation (ALDH2*2) involves an E to K (E478K) substitution in the catalytic domain that leads to decreased enzymatic activity.Scientists hypothesize that Asian populations have higher rates of ALDH2*2 incidence. The properties of ALDH2 variants in Caucasian and Japanese populations were analyzed in the following experiments.Experiment 1Mitochondrial extracts from both populations were combined in a single mixture and separated by native gel electrophoresis. Proteins 210-230 kD in size were removed from the agarose gel and further isolated using affinity columns with antibodies designed to separate wild-type ALDH2 from ALDH2*2 by binding the catalytic domain of the wild-type but not the mutant protein. Concentrated eluates E1 and E2 were produced by washing the affinity columns with buffers that remove unbound proteins and dislodge bound proteins from antibodies, respectively (Figure 1) .
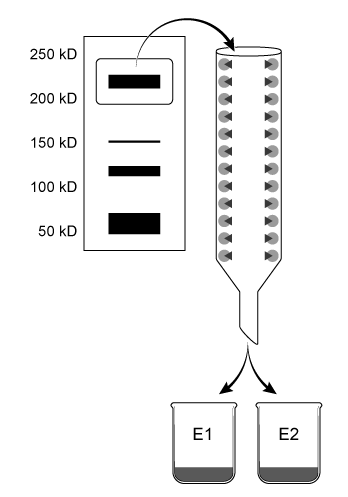 Figure 1 Purification of ALDH2 proteinsALDH2 activity in each eluate was quantified by adding acetaldehyde and NAD+, and then monitoring the rate of NADH production. The results are shown in Figure 2.
Figure 1 Purification of ALDH2 proteinsALDH2 activity in each eluate was quantified by adding acetaldehyde and NAD+, and then monitoring the rate of NADH production. The results are shown in Figure 2.
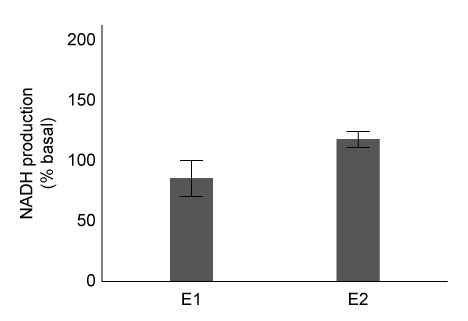 Figure 2 Enzymatic activity of E1 and E2 eluatesExperiment 2Researchers predict that mutations at conserved amino acid sites containing S or T may prevent ALDH2 from being phosphorylated by protein kinase C epsilon (εPKC) . The activity of wild-type ALDH2 (WT) and ALDH2 mutants with substitutions at positions T185 and S279 in the presence or absence of εPKC are summarized in Table 1.Table 1 Effect of εPKC on ALDH2 Variants
Figure 2 Enzymatic activity of E1 and E2 eluatesExperiment 2Researchers predict that mutations at conserved amino acid sites containing S or T may prevent ALDH2 from being phosphorylated by protein kinase C epsilon (εPKC) . The activity of wild-type ALDH2 (WT) and ALDH2 mutants with substitutions at positions T185 and S279 in the presence or absence of εPKC are summarized in Table 1.Table 1 Effect of εPKC on ALDH2 Variants
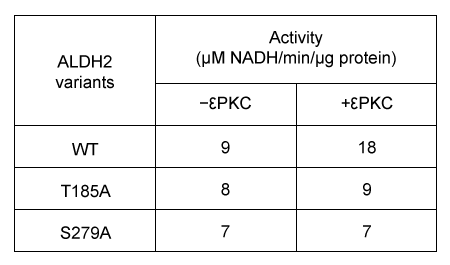 Adapted from Chen CH, Ferreira JC, Gross ER, Mochly-rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014;94(1) :1-34.
-Scientists could confirm that Asian populations have higher rates of ALDH2*2 mutations if they discovered which of the following?
Adapted from Chen CH, Ferreira JC, Gross ER, Mochly-rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014;94(1) :1-34.
-Scientists could confirm that Asian populations have higher rates of ALDH2*2 mutations if they discovered which of the following?
A) The proteins in E2 were exclusively from Asian samples.
B) None of the proteins in E1 were from Caucasian samples.
C) Only WT-ALDH2 proteins were eluted from the affinity column.
D) ALDH2*2 proteins were eluted first from the affinity column.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Destabilase, an enzyme found in the saliva of leeches, exhibits at least two activities: isopeptidase activity and glycosidase activity. Isopeptidase activity is believed to prevent blood clotting by hydrolyzing the isopeptide bond between the ε-amino group of lysine and the γ-carboxyl group of glutamate in fibrin monomers. The enzyme's glycosidase activity digests sugars found in the peptidoglycans of bacterial cell walls and may play a role in innate immunity.Two isoforms of destabilase were tagged with a polyhistidine tail and recombinantly expressed in E. coli cells. Both isoforms were then purified by affinity chromatography and eluted by imidazole, which competes with histidine-tagged proteins for column binding. Each isoform eluted efficiently with 300 mM imidazole. Five microliters of each purified enzyme were then provided with saturating levels of substrate and assessed for isopeptidase and glycosidase activity at various pH levels, which allowed the optimal pH of enzymatic activities for each isoform to be identified (Figure 1) . Further studies showed that at optimal pH, both isoforms had approximately the same kcat values, but isoform 2 had a lower Km than isoform 1 for both enzymatic activities.
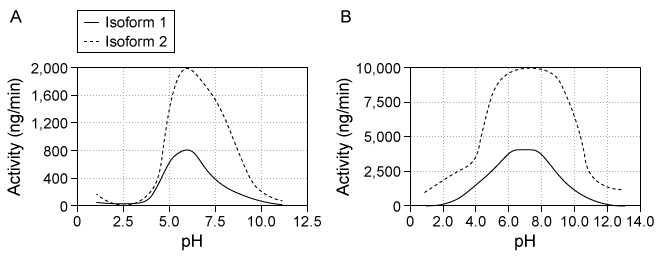 Figure 1 Isopeptidase activity (A) and glycosidase activity (B) of 5 μL of destabilase isoforms 1 and 2 at various pH valuesIsoform 2 was further characterized by mutating specific residues and measuring enzymatic activities at optimal pH. The results are shown in Figure 2.
Figure 1 Isopeptidase activity (A) and glycosidase activity (B) of 5 μL of destabilase isoforms 1 and 2 at various pH valuesIsoform 2 was further characterized by mutating specific residues and measuring enzymatic activities at optimal pH. The results are shown in Figure 2.
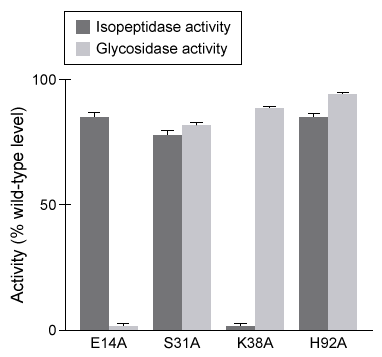 Figure 2 Isopeptidase and glycosidase activity of isoform 2 of destabilase with various mutations
-Given that blood pH is 7.4, what is the approximate isopeptidase activity level of isoform 1 in blood expressed as a percentage of optimal activity?
Figure 2 Isopeptidase and glycosidase activity of isoform 2 of destabilase with various mutations
-Given that blood pH is 7.4, what is the approximate isopeptidase activity level of isoform 1 in blood expressed as a percentage of optimal activity?
A) 40%
B) 50%
C) 80%
D) 100%
Correct Answer

verified
Correct Answer
verified
Multiple Choice
The Km of an enzyme is always decreased by which class of inhibitor?
A) Mixed
B) Noncompetitive
C) Competitive
D) Uncompetitive
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Mitochondria are double membrane-bound cellular organelles that house several connected metabolic reactions. The citric acid cycle takes place inside the mitochondrial matrix and produces two reduced electron carriers, NADH and FADH2. These two carriers then pass their electrons to ubiquinone (UQ) , which is reduced to ubiquinol (UQH2) . UQH2 passes its electrons to oxidized cytochrome C (cyt-Cox) , regenerating UQ and forming reduced cytochrome C (cyt-Cred) . Finally, cyt-Cred transfers electrons to oxygen, regenerating cyt-Cox and forming water. Each reaction is facilitated by one of four protein complexes that are collectively known as the electron transport chain (ETC) .Researchers isolated mitochondria and treated them with a novel ETC inhibitor, "drug X." After an overnight incubation, they measured levels of several ETC-reduced electron carriers in the presence of drug X relative to untreated mitochondria. Their results are shown in Figure 1.
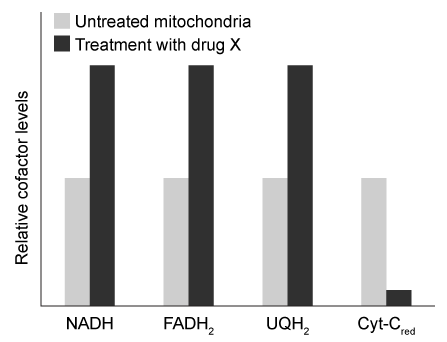 Figure 1 Relative levels of reduced electron carriers in the presence and absence of drug XNext, the researchers exposed mitochondria to FCCP, a chemical that can transport protons across membranes. They observed that in the presence of FCCP, the citric acid cycle was fully active, and all reduced cofactors were present at the same levels as in untreated mitochondria. However, the rate of ATP synthesis was significantly reduced. ATP synthesis was immediately restored when FCCP was washed away.
-Based on the information in the passage, FCCP most likely causes a decrease in energy production by:
Figure 1 Relative levels of reduced electron carriers in the presence and absence of drug XNext, the researchers exposed mitochondria to FCCP, a chemical that can transport protons across membranes. They observed that in the presence of FCCP, the citric acid cycle was fully active, and all reduced cofactors were present at the same levels as in untreated mitochondria. However, the rate of ATP synthesis was significantly reduced. ATP synthesis was immediately restored when FCCP was washed away.
-Based on the information in the passage, FCCP most likely causes a decrease in energy production by:
A) decoupling the movement of protons down their concentration gradient from ATP synthase.
B) carrying more protons into the intermembrane space than are pumped by the ETC alone.
C) shifting the proton gradient further away from equilibrium than is achieved by the ETC alone.
D) causing a decrease in ETC activity, resulting in fewer available protons for ATP synthase.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
DNA polymerization is one of the most conserved mechanisms of genome replication. Synthesis of a complete DNA strand requires a template, primers, a polymerase enzyme, and sufficient deoxyribonucleotide triphosphates (dNTPs) . The DNA polymerase enzyme binds consecutive base pairs on the template strand and extends the double helix by adding dNTPs to the primer. The amino acid residues in the active site of DNA polymerase form hydrogen bonds with Watson-Crick donors and acceptors on incoming DNA nucleotides to facilitate base pairing.The formation of the DNA double helix creates opposing changes in entropy and enthalpy. Favorable bonding interactions via hydrogen bonds during Watson-Crick base pairing results in negative enthalpy, and restricted rotation and flexibility of the DNA backbone generates negative entropy. Scientists hypothesize that hydrogen bonding between bases not only stabilizes the double helix but is also crucial for selective and efficient replication.Analogs that are similar in size and shape to naturally occurring bases can be used to determine the influence of hydrogen bonding on base pair selectivity. To mimic the structure of deoxythymidine triphosphate (dTTP) , researchers synthesized dNTP derivatives of difluorotoluene (dFTP) , a nonpolar analog that lacks Watson-Crick hydrogen bonding. Klenow fragment (KF) polymerase, which has 3′-5′ but not 5′-3′ exonuclease activity, was incubated with a mixture of DNA template, primers, and dNTPs, including dFTP derivatives. The efficiency of dFTP and natural dTTP nucleotide incorporation into a growing primer strand by KF is shown in Figure 1.
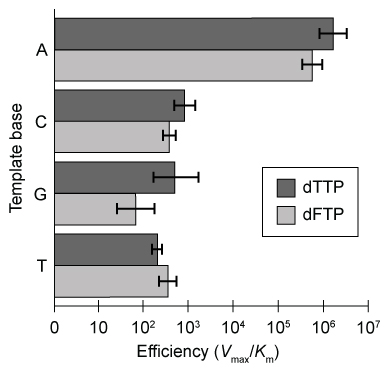 Figure 1 Template-specific selection of dFTP and dTTP by the KF enzyme
Adapted from Moran S, Ren RX, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc Natl Acad Sci USA. 1997;94(20) :10506-11.
-Based on the information in the passage, what effect would incorporation of dFTP in place of dTTP most likely have on the melting temperature of a DNA molecule of a fixed length?
Figure 1 Template-specific selection of dFTP and dTTP by the KF enzyme
Adapted from Moran S, Ren RX, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc Natl Acad Sci USA. 1997;94(20) :10506-11.
-Based on the information in the passage, what effect would incorporation of dFTP in place of dTTP most likely have on the melting temperature of a DNA molecule of a fixed length?
A) The melting temperature would increase due to reduced base stacking.
B) The melting temperature would decrease due to decreased hydrogen bonding.
C) The melting temperature would not change because dFTP is similar to dTTP.
D) The effect on melting temperature depends on the number of nucleotides in the DNA strand.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
The innate immune system relies heavily on the phagocytic action of neutrophils, mobile white blood cells that engulf and degrade bacteria and viruses as the first line of host defense against invading pathogens. One mechanism for eliminating viruses engulfed in the phagosome involves the degradation of viral proteins by neutrophil serine proteases (NSPs) .NSPs, which form part of the chymotrypsin family of serine proteases, cleave viral proteins using a catalytic triad relay system. Originally discovered in granules released by neutrophils, NSP4 is a novel monomeric serine protease that has been studied to elucidate the catalytic mechanism of NSPs. Prior experimental data suggest that NSP4 alters its tertiary structure upon substrate binding and cleaves viral proteins using the acylation reaction shown in Figure 1.
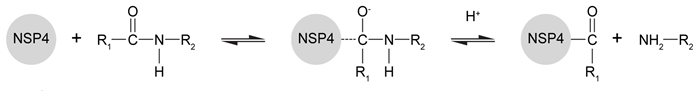 Figure 1 Degradation of viral proteins by acylation of NSP4Computational methods were used to identify the energetic parameters of reactions between NSP4 and synthetic viral proteins. To ensure that simulated reactions resembled physiological conditions, algorithms were adjusted to account for the enzyme being in an aqueous environment. The rate-determining step in the acylation reaction was determined by calculating the changes in free energy in both the enzyme and substrate (Figure 2) .
Figure 1 Degradation of viral proteins by acylation of NSP4Computational methods were used to identify the energetic parameters of reactions between NSP4 and synthetic viral proteins. To ensure that simulated reactions resembled physiological conditions, algorithms were adjusted to account for the enzyme being in an aqueous environment. The rate-determining step in the acylation reaction was determined by calculating the changes in free energy in both the enzyme and substrate (Figure 2) .
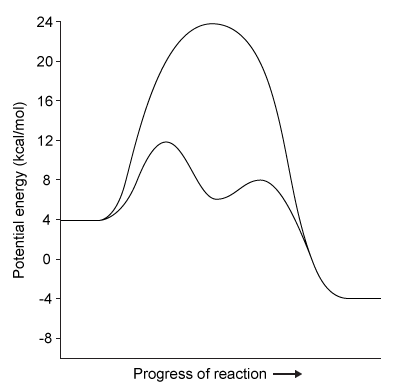 Figure 2 Free energy profiles of synthetic viral protein cleavageKnowledge of amino acid frequency distribution at an enzymatic active site may facilitate the design of novel active sites and enzyme-specific inhibitors. Researchers analyzed the composition of amino acids that form the NSP4 active site during protein folding to study the characteristics of residues involved in active site formation (Figure 3) .
Figure 2 Free energy profiles of synthetic viral protein cleavageKnowledge of amino acid frequency distribution at an enzymatic active site may facilitate the design of novel active sites and enzyme-specific inhibitors. Researchers analyzed the composition of amino acids that form the NSP4 active site during protein folding to study the characteristics of residues involved in active site formation (Figure 3) .
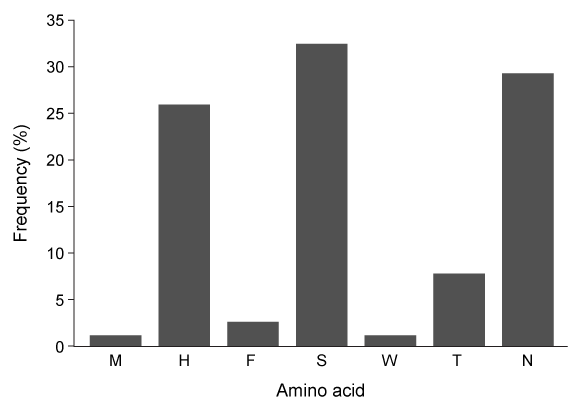 Figure 3 Frequency distribution of amino acid residues at the NSP4 active site
Adapted from Stapels DA, Geisbrecht BV, Rooijakkers SH. Neutrophil serine proteases in antibacterial defense. Curr Opin Microbiol. 2015;23:42-8.
-Based on the passage, the cleavage of viral proteins by NSP4 is:
Figure 3 Frequency distribution of amino acid residues at the NSP4 active site
Adapted from Stapels DA, Geisbrecht BV, Rooijakkers SH. Neutrophil serine proteases in antibacterial defense. Curr Opin Microbiol. 2015;23:42-8.
-Based on the passage, the cleavage of viral proteins by NSP4 is:
A) an exergonic process moving in the reverse direction.
B) an endergonic process moving in the forward direction.
C) a nonspontaneous reaction requiring energy to proceed.
D) a spontaneous reaction resulting in a release of energy.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
NDPK is an enzyme in budding yeast that uses ATP to phosphorylate nucleotide diphosphates (NDPs and dNDPs) , yielding the triphosphate form of each (NTPs and dNTPs) . dNTPs are necessary for DNA synthesis, yet yeast cells that lack NDPK grow and divide at the same rate as wild-type yeast. Therefore, triphosphate generation is most likely carried out by one or more other enzymes.The enzyme TMPK converts deoxythymidine monophosphate (dTMP) to the diphosphate form (dTDP) by transferring the γ-phosphate of ATP to dTMP. Yeast cells that lack TMPK activity cannot synthesize DNA and become arrested during replication. It was proposed that TMPK may partially compensate for NDPK loss by phosphorylating dTDP as well as dTMP.To better understand the role of TMPK in nucleotide phosphorylation, wild-type TMPK and the inactive E75K variant were purified by affinity chromatography, and the following experiments were performed.Experiment 1After purification, a sample containing 3 μg of protein was added to a reaction mixture containing radiolabeled nucleotide monophosphates (NMPs or dNMPs) and unlabeled ATP. The reaction proceeded for 30 minutes, and products were separated by thin-layer chromatography (TLC) on a PEI-cellulose plate. PEI-cellulose separates nucleotides by charge (Figure 1) , and migration distance decreases with increasing negative charge. Spots were visualized by autoradiography, and it was determined that 0.30 nmol of dTMP and 0.15 nmol of uridine monophosphate (UMP) became phosphorylated, respectively.
 Figure 1 TLC analysis of nucleotide monophosphates after exposure to TMPK and ATP. (Note: Different nucleotides with the same phosphorylation level migrate different distances.) Experiment 2The experiment was repeated using radiolabeled dTDP as the starting material. The reaction proceeded in buffer only, in the presence of E75K TMPK, or in the presence of wild-type TMPK. Products were separated by TLC and visualized (Figure 2) .
Figure 1 TLC analysis of nucleotide monophosphates after exposure to TMPK and ATP. (Note: Different nucleotides with the same phosphorylation level migrate different distances.) Experiment 2The experiment was repeated using radiolabeled dTDP as the starting material. The reaction proceeded in buffer only, in the presence of E75K TMPK, or in the presence of wild-type TMPK. Products were separated by TLC and visualized (Figure 2) .
 Figure 2 TLC analysis of dTDP phosphorylation with buffer only (Lane 1) , E75K TMPK (Lane 2) , or wild-type TMPK (Lane 3)
Adapted from Chien CY, Chen BR, Chou CK, Sclafani RA, Su JY. The yeast Cdc8 exhibits both deoxythymidine monophosphate and diphosphate kinase activities. FEBS Lett. 2009;583(13) :2281-6.
-Experiment 2 was performed to determine whether phosphorylation of dTDP was due to residual NDPK activity in the purified enzyme solution. Which of the following shows the expected outcome of Experiment 2 if NDPK were still present after purification?
Figure 2 TLC analysis of dTDP phosphorylation with buffer only (Lane 1) , E75K TMPK (Lane 2) , or wild-type TMPK (Lane 3)
Adapted from Chien CY, Chen BR, Chou CK, Sclafani RA, Su JY. The yeast Cdc8 exhibits both deoxythymidine monophosphate and diphosphate kinase activities. FEBS Lett. 2009;583(13) :2281-6.
-Experiment 2 was performed to determine whether phosphorylation of dTDP was due to residual NDPK activity in the purified enzyme solution. Which of the following shows the expected outcome of Experiment 2 if NDPK were still present after purification?
A) ![]()
B) ![]()
C) ![]()
D) ![]()
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Which of the following peptides would exhibit the greatest steric constraints?
A) AGSCTN
B) ICGHLA
C) PDNETQ
D) FYLWIR
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
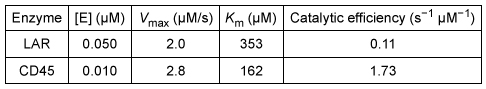
Leukocyte common antigen (LCA) enzymes play several important developmental roles in both vertebrates and invertebrates. They regulate numerous cellular processes by catalyzing Reaction 1 in target proteins at specific developmental stages. Consequently, LCA enzyme expression and activity are tightly controlled in various cell types.
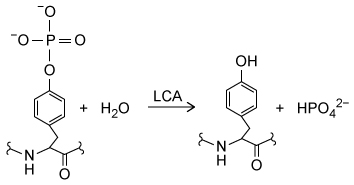 Reaction 1The Src protein is critical for cell differentiation and motility, and is activated by LCA enzyme activity. Two LCA enzymes, known as LAR and CD45, were purified and incubated with the phosphopeptide src527 (Figure 1) , derived from Src.
Reaction 1The Src protein is critical for cell differentiation and motility, and is activated by LCA enzyme activity. Two LCA enzymes, known as LAR and CD45, were purified and incubated with the phosphopeptide src527 (Figure 1) , derived from Src.
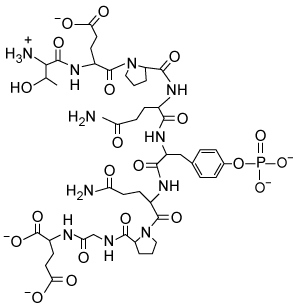 Figure 1 Structure of the src527 peptide used as a substrate in LAR and CD45 assaysThe rate of Reaction 1 was measured at various concentrations of src527 in the presence of either LAR or CD45, and kinetic parameters were determined (Table 1) .Table 1 Kinetic Parameters of LAR and CD45
Figure 1 Structure of the src527 peptide used as a substrate in LAR and CD45 assaysThe rate of Reaction 1 was measured at various concentrations of src527 in the presence of either LAR or CD45, and kinetic parameters were determined (Table 1) .Table 1 Kinetic Parameters of LAR and CD45
 Compound 1 was synthesized and tested for its effectiveness as an inhibitor of LCA enzymes.
Compound 1 was synthesized and tested for its effectiveness as an inhibitor of LCA enzymes.
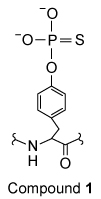 The study in Table 1 was repeated in the presence of Compound 1, which was found to significantly reduce the activities of both LAR and CD45 against the src527 peptide. Compound 1 was subsequently used in assays to examine the in vivo function of LCA enzymes.
Adapted from Cho H, Krishnaraj R, Itoh M, et al. Substrate specificities of catalytic fragments of protein tyrosine phosphatases (HPTP beta, LAR, and CD45) toward phosphotyrosylpeptide substrates and thiophosphotyrosylated peptides as inhibitors. Protein Sci. 1993;2(6) :977-84.
-Compound 1 would most likely have what effect on the activity of LAR?
The study in Table 1 was repeated in the presence of Compound 1, which was found to significantly reduce the activities of both LAR and CD45 against the src527 peptide. Compound 1 was subsequently used in assays to examine the in vivo function of LCA enzymes.
Adapted from Cho H, Krishnaraj R, Itoh M, et al. Substrate specificities of catalytic fragments of protein tyrosine phosphatases (HPTP beta, LAR, and CD45) toward phosphotyrosylpeptide substrates and thiophosphotyrosylated peptides as inhibitors. Protein Sci. 1993;2(6) :977-84.
-Compound 1 would most likely have what effect on the activity of LAR?
A) It would bind to an allosteric site and decrease the Km.
B) It would bind to the active site and decrease the Vmax.
C) It would bind to an allosteric site and increase the Vmax.
D) It would bind to the active site and increase the Km.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Myalgic encephalopathy (ME) , or chronic fatigue syndrome (CFS) , is a physiological disease that affects 0.15% of the human population. Compared with healthy individuals, persons with ME experience reduced phospholipid levels, muscular degeneration, urinary urgency, and other symptoms that contribute to a lower quality of life. Studies in resting subjects have yielded inconsistent data and failed to identify a unique metabolic defect in patients with ME.Analyzing the blood plasma of male and female patients with ME, physicians have observed that male patients display elevated levels of 3-methylhistidine, a marker of protein catabolism. Researchers expanded this finding by measuring the serum concentration of amino acids in blood samples obtained from nonfasting ME patients and healthy controls. Amino acids were classified into separate categories according to their metabolic intermediates.Table 1 Classification of Amino Acids in Blood Plasma
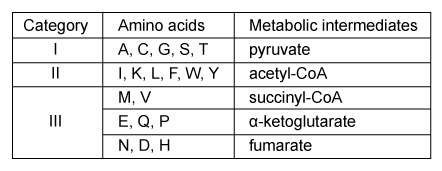
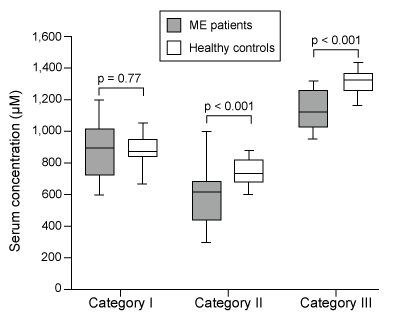 Figure 1 Serum concentrations of amino acids in patients with ME and healthy controls (HC) Next, researchers hypothesized that the inhibition of pyruvate dehydrogenase (PDH) and impairment of fatty acid oxidation were responsible for the fluctuations in amino acid levels. PDH function and lipid metabolism were studied in peripheral blood mononuclear cells (PBMCs) by measuring the gene expression of proteins within regulatory pathways. Researchers monitored the mRNA levels of the inhibitor pyruvate dehydrogenase kinase 1 (PDK1) and of sirtuin 4 (SIRT4) , which removes lipoic acid groups from the PDH complex. They also examined lipid metabolism by measuring the mRNA levels of peroxisome proliferator-activated receptor-α (PPARα) , a transcription factor that upregulates the uptake of fatty acids. The mRNA level of each protein in PBMCs of patients with ME was evaluated against healthy controls, as shown in Figure 2.
Figure 1 Serum concentrations of amino acids in patients with ME and healthy controls (HC) Next, researchers hypothesized that the inhibition of pyruvate dehydrogenase (PDH) and impairment of fatty acid oxidation were responsible for the fluctuations in amino acid levels. PDH function and lipid metabolism were studied in peripheral blood mononuclear cells (PBMCs) by measuring the gene expression of proteins within regulatory pathways. Researchers monitored the mRNA levels of the inhibitor pyruvate dehydrogenase kinase 1 (PDK1) and of sirtuin 4 (SIRT4) , which removes lipoic acid groups from the PDH complex. They also examined lipid metabolism by measuring the mRNA levels of peroxisome proliferator-activated receptor-α (PPARα) , a transcription factor that upregulates the uptake of fatty acids. The mRNA level of each protein in PBMCs of patients with ME was evaluated against healthy controls, as shown in Figure 2.
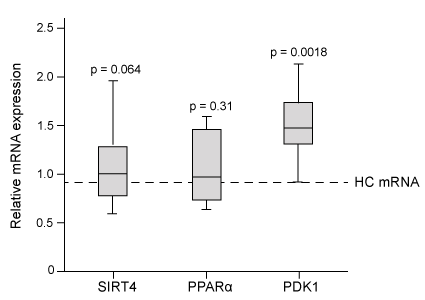 Figure 2 Gene expression of PDK1, SIRT4, and PPARα in PBMCs
Adapted from Fluge Ø, Mella O, Bruland O, et al.. JCI Insight. 2016;1(21) :e89376.
-To confirm that β-oxidation is NOT impaired in healthy controls, researchers most likely measured which metabolic step in PBMCs?
Figure 2 Gene expression of PDK1, SIRT4, and PPARα in PBMCs
Adapted from Fluge Ø, Mella O, Bruland O, et al.. JCI Insight. 2016;1(21) :e89376.
-To confirm that β-oxidation is NOT impaired in healthy controls, researchers most likely measured which metabolic step in PBMCs?
A) The exportation of citrate to the cytoplasm to produce acetyl-CoA
B) The release of CO2 from malonyl-CoA
C) The translocation of acylcarnitine to the mitochondrial matrix
D) The depletion of reduced electron carriers
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate.
![Passage Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate. <strong>Figure 1</strong> AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.<strong>Table 1</strong> ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43. -The structure of Compound 1, which is found in adipocytes, is shown below. What would be the catabolic fate of this molecule? A) Part A enters glycolysis and part B enters β-oxidation. B) Part A enters gluconeogenesis and part B enters fatty acid synthesis. C) Part B enters fatty acid synthesis and part A enters β-oxidation. D) Part B enters glycogenolysis and part A enters glycolysis.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0dc2_af71_a3bf_719de41c80b7_MD0008_00.jpg) Figure 1 AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.Table 1 ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo
Figure 1 AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.Table 1 ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo
![Passage Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate. <strong>Figure 1</strong> AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.<strong>Table 1</strong> ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43. -The structure of Compound 1, which is found in adipocytes, is shown below. What would be the catabolic fate of this molecule? A) Part A enters glycolysis and part B enters β-oxidation. B) Part A enters gluconeogenesis and part B enters fatty acid synthesis. C) Part B enters fatty acid synthesis and part A enters β-oxidation. D) Part B enters glycogenolysis and part A enters glycolysis.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0dc2_af72_a3bf_e9a0f8dccdf8_MD0008_00.jpg) Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43.
-The structure of Compound 1, which is found in adipocytes, is shown below. What would be the catabolic fate of this molecule?
Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43.
-The structure of Compound 1, which is found in adipocytes, is shown below. What would be the catabolic fate of this molecule? ![Passage Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate. <strong>Figure 1</strong> AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.<strong>Table 1</strong> ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43. -The structure of Compound 1, which is found in adipocytes, is shown below. What would be the catabolic fate of this molecule? A) Part A enters glycolysis and part B enters β-oxidation. B) Part A enters gluconeogenesis and part B enters fatty acid synthesis. C) Part B enters fatty acid synthesis and part A enters β-oxidation. D) Part B enters glycogenolysis and part A enters glycolysis.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0dc2_d683_a3bf_7db95eb731ca_MD0008_00.jpg)
A) Part A enters glycolysis and part B enters β-oxidation.
B) Part A enters gluconeogenesis and part B enters fatty acid synthesis.
C) Part B enters fatty acid synthesis and part A enters β-oxidation.
D) Part B enters glycogenolysis and part A enters glycolysis.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
In the electron transport chain, four electrons are required to fully reduce one O2 molecule to two H2O molecules. Given this information, how many NADH molecules are required?
A) 1
B) 2
C) 3
D) 4
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Physical activity produces an increased energy requirement in the muscles as well as in the brain. During periods of intense exercise, glycogenolysis is activated in muscles to provide them with the energy needed for contractions, and research indicates that astrocytes also consume glycogen at increased levels during exercise. It is hypothesized that during periods of physical exertion, astrocytes convert glycogen to lactate, which is then used as a fuel source in neurons.Monocarboxylate transporters (MCTs) are responsible for the movement of lactate across membranes, and MCT2 is expressed at increased levels in neuronal cells during exercise. Scientists proposed that lactate is secreted from astrocytes and taken up by neurons through MCT2. Lactate is then converted to pyruvate and enters the mitochondria, where it is further metabolized. To test their predictions, scientists examined the effect of exercise on glycogen metabolism in rodents.Experiment 1Rats were either allowed to rest or exercised to exhaustion. The levels of several metabolites were then quantified in both the plantaris muscle and the hippocampus.
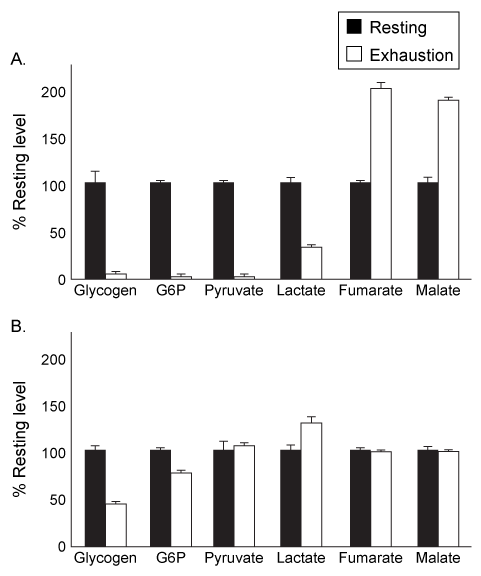 Figure 1 Levels of several metabolites in plantaris muscle (A) and hippocampus (B) from rats at rest or after exercising to exhaustionExperiment 2Rats were injected with 1,4-dideoxy-1,4-imino-D-arabinitol (DAB) or saline, and glycogen and lactate levels were measured in the hippocampus of rats at rest or after exercise to exhaustion.
Figure 1 Levels of several metabolites in plantaris muscle (A) and hippocampus (B) from rats at rest or after exercising to exhaustionExperiment 2Rats were injected with 1,4-dideoxy-1,4-imino-D-arabinitol (DAB) or saline, and glycogen and lactate levels were measured in the hippocampus of rats at rest or after exercise to exhaustion.
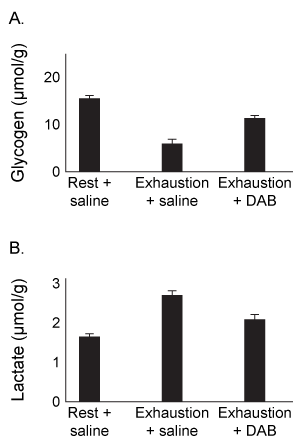 Figure 2 Levels of glycogen (A) and lactate (B) in rats injected with DAB or saline, after rest or upon exhaustion
Adapted from Matsui T, Omuro H, Liu Y-F, et al. Astrocytic glycogen-derived lactate fuels the brain during exhaustive exercise to maintain endurance capacity. Proceedings of the National Academy of Sciences. 2017;114(24) :6358-6363.
-If the proposed mechanism in the passage is correct, which of the following metabolic reactions must occur for astrocytes to convert glycogen into a fuel for neurons?Glucose-1-phosphate to glucose-6-phosphateGlucose to glucose-6-phosphate3-phosphoglycerate to 2-phosphoglyceratePyruvate to acetyl-CoAPyruvate to lactate
Figure 2 Levels of glycogen (A) and lactate (B) in rats injected with DAB or saline, after rest or upon exhaustion
Adapted from Matsui T, Omuro H, Liu Y-F, et al. Astrocytic glycogen-derived lactate fuels the brain during exhaustive exercise to maintain endurance capacity. Proceedings of the National Academy of Sciences. 2017;114(24) :6358-6363.
-If the proposed mechanism in the passage is correct, which of the following metabolic reactions must occur for astrocytes to convert glycogen into a fuel for neurons?Glucose-1-phosphate to glucose-6-phosphateGlucose to glucose-6-phosphate3-phosphoglycerate to 2-phosphoglyceratePyruvate to acetyl-CoAPyruvate to lactate
A) I, II, and III only
B) II, III, and V only
C) I, IV, and V only
D) I, III, and V only
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Nerve cells must maintain the composition of their myelin sheaths to function properly. Galactocerebroside (GC) , shown in Figure 1, is a hydrolyzable lipid and an important component of the myelin sheath.
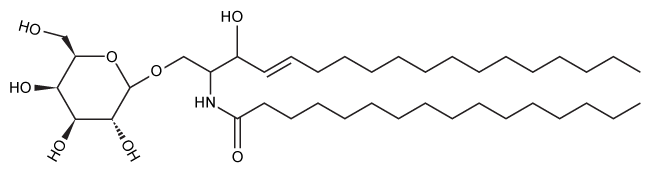 Figure 1 Structure of the myelin sheath component galactocerebrosideHomeostasis of GC is maintained by its dynamic synthesis and degradation, as shown in Figure 2. GC is synthesized by the enzyme ceramide galactosyltransferase (CGT) using UDP-galactose (UDP-Gal) and ceramide (Cer) as substrates. It is degraded by the enzyme galactocerebrosidase (GalC) , which hydrolyzes the glycosidic bond to release galactose (Gal) and ceramide. A deficiency in GalC activity leads to a buildup of GC, and prior research has connected this buildup with neurodegeneration.
Figure 1 Structure of the myelin sheath component galactocerebrosideHomeostasis of GC is maintained by its dynamic synthesis and degradation, as shown in Figure 2. GC is synthesized by the enzyme ceramide galactosyltransferase (CGT) using UDP-galactose (UDP-Gal) and ceramide (Cer) as substrates. It is degraded by the enzyme galactocerebrosidase (GalC) , which hydrolyzes the glycosidic bond to release galactose (Gal) and ceramide. A deficiency in GalC activity leads to a buildup of GC, and prior research has connected this buildup with neurodegeneration.
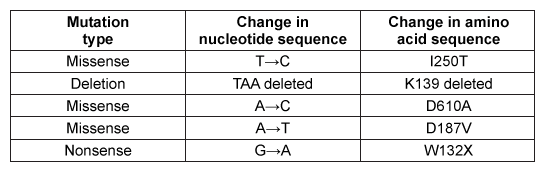
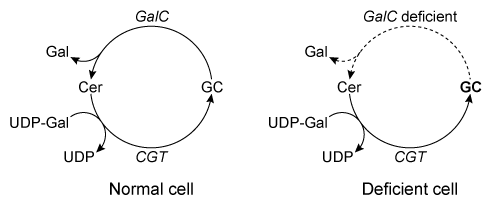 Figure 2 Schematic of GCT and GalC activity in healthy and GalC deficient cellsOver 100 inactivating GalC mutations have been identified. Five of the most common mutations are listed in Table 1. The changes to both DNA and amino acid sequence are shown.Table 1 Five Common Galactocerebrosidase Mutations
Figure 2 Schematic of GCT and GalC activity in healthy and GalC deficient cellsOver 100 inactivating GalC mutations have been identified. Five of the most common mutations are listed in Table 1. The changes to both DNA and amino acid sequence are shown.Table 1 Five Common Galactocerebrosidase Mutations
 -GC can be classified as:
-GC can be classified as:
A) a glycerophospholipid.
B) a phosphosphingolipid.
C) a glycosphingolipid.
D) a phosphoglycolipid.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Diabetic nephropathy (kidney disease) is characterized in part by a significant decrease in the glomerular filtration rate. Podocytes are differentiated cells that surround the glomerular capillaries and counteract the pressure of blood filtration. Hyperglycemia (elevated blood glucose) and capillary hypertension (high blood pressure) , both of which are common in diabetic patients, can subject podocytes to mechanical stress. This mechanical stress has been shown to regulate glucose uptake by podocytes, possibly by altering the production of glucose transport (GLUT) proteins.Researchers investigating GLUT2 and GLUT4 expression in rat podocytes measured the impact of mechanical stress on glucose uptake. Cells were grown for 4 hours on membranes exposed to atmospheric (atm) or high air pressure (HP) to mimic mechanical stress. Antibodies specific to GLUT2 and GLUT4 were used to visualize the expression of each under normal glucose (NG) and hyperglycemic (HG) conditions, and to assess the effect of mechanical stress on expression. The uptake of a radiolabeled glucose analogue, [3H]-2-deoxy-D-glucose (3H-2DG) , was then determined. The quantity of 3H-2DG transported by each GLUT protein was directly proportional to the cell surface expression.
![Passage Diabetic nephropathy (kidney disease) is characterized in part by a significant decrease in the glomerular filtration rate. Podocytes are differentiated cells that surround the glomerular capillaries and counteract the pressure of blood filtration. Hyperglycemia (elevated blood glucose) and capillary hypertension (high blood pressure) , both of which are common in diabetic patients, can subject podocytes to mechanical stress. This mechanical stress has been shown to regulate glucose uptake by podocytes, possibly by altering the production of glucose transport (GLUT) proteins.Researchers investigating GLUT2 and GLUT4 expression in rat podocytes measured the impact of mechanical stress on glucose uptake. Cells were grown for 4 hours on membranes exposed to atmospheric (atm) or high air pressure (HP) to mimic mechanical stress. Antibodies specific to GLUT2 and GLUT4 were used to visualize the expression of each under normal glucose (NG) and hyperglycemic (HG) conditions, and to assess the effect of mechanical stress on expression. The uptake of a radiolabeled glucose analogue, [<sup>3</sup>H]-2-deoxy-D-glucose (3H-2DG) , was then determined. The quantity of 3H-2DG transported by each GLUT protein was directly proportional to the cell surface expression. <strong>Figure 1</strong> Cell surface expression of GLUT transporters in response to mechanical stress and varying glucose concentration.In vivo studies have shown that transport via GLUT2 is noninducible and depends only on extracellular glucose concentrations. However, GLUT4-mediated uptake is induced by insulin binding and occurs under high glucose conditions when insulin is released. The standard K<sub>m</sub> values for GLUT2 and GLUT4 are 19.4 and 4.3 mM, respectively, at normal blood glucose levels after a meal (~7.8 mM) . The data shown in Figure 2 represent the combined kinetic properties of both transporters. <strong>Figure 2</strong> Plot of glucose uptake in podocytes with and without mechanical stress. Adapted from Lewko, B., Bryl, E., Witkowski, J. M., Latawiec, E., Angielski, S., & Stepinski, J. (2005) . Mechanical stress and glucose concentration modulate glucose transport in cultured rat podocytes. Nephrology Dialysis Transplantation, 20(2) , 306-311. -Which statement about glucose transport under physiological conditions is consistent with the kinetic data provided in the passage? A) GLUT2-mediated uptake is a first-order process. B) ATP is required for glucose uptake by both GLUT transporters. C) In the fed state, glucose concentration limits GLUT4-mediated uptake. D) Glucose transport is independent of the concentration gradient.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0e7c_899d_a3bf_e372713321b8_MD0008_00.jpg) Figure 1 Cell surface expression of GLUT transporters in response to mechanical stress and varying glucose concentration.In vivo studies have shown that transport via GLUT2 is noninducible and depends only on extracellular glucose concentrations. However, GLUT4-mediated uptake is induced by insulin binding and occurs under high glucose conditions when insulin is released. The standard Km values for GLUT2 and GLUT4 are 19.4 and 4.3 mM, respectively, at normal blood glucose levels after a meal (~7.8 mM) . The data shown in Figure 2 represent the combined kinetic properties of both transporters.
Figure 1 Cell surface expression of GLUT transporters in response to mechanical stress and varying glucose concentration.In vivo studies have shown that transport via GLUT2 is noninducible and depends only on extracellular glucose concentrations. However, GLUT4-mediated uptake is induced by insulin binding and occurs under high glucose conditions when insulin is released. The standard Km values for GLUT2 and GLUT4 are 19.4 and 4.3 mM, respectively, at normal blood glucose levels after a meal (~7.8 mM) . The data shown in Figure 2 represent the combined kinetic properties of both transporters.
![Passage Diabetic nephropathy (kidney disease) is characterized in part by a significant decrease in the glomerular filtration rate. Podocytes are differentiated cells that surround the glomerular capillaries and counteract the pressure of blood filtration. Hyperglycemia (elevated blood glucose) and capillary hypertension (high blood pressure) , both of which are common in diabetic patients, can subject podocytes to mechanical stress. This mechanical stress has been shown to regulate glucose uptake by podocytes, possibly by altering the production of glucose transport (GLUT) proteins.Researchers investigating GLUT2 and GLUT4 expression in rat podocytes measured the impact of mechanical stress on glucose uptake. Cells were grown for 4 hours on membranes exposed to atmospheric (atm) or high air pressure (HP) to mimic mechanical stress. Antibodies specific to GLUT2 and GLUT4 were used to visualize the expression of each under normal glucose (NG) and hyperglycemic (HG) conditions, and to assess the effect of mechanical stress on expression. The uptake of a radiolabeled glucose analogue, [<sup>3</sup>H]-2-deoxy-D-glucose (3H-2DG) , was then determined. The quantity of 3H-2DG transported by each GLUT protein was directly proportional to the cell surface expression. <strong>Figure 1</strong> Cell surface expression of GLUT transporters in response to mechanical stress and varying glucose concentration.In vivo studies have shown that transport via GLUT2 is noninducible and depends only on extracellular glucose concentrations. However, GLUT4-mediated uptake is induced by insulin binding and occurs under high glucose conditions when insulin is released. The standard K<sub>m</sub> values for GLUT2 and GLUT4 are 19.4 and 4.3 mM, respectively, at normal blood glucose levels after a meal (~7.8 mM) . The data shown in Figure 2 represent the combined kinetic properties of both transporters. <strong>Figure 2</strong> Plot of glucose uptake in podocytes with and without mechanical stress. Adapted from Lewko, B., Bryl, E., Witkowski, J. M., Latawiec, E., Angielski, S., & Stepinski, J. (2005) . Mechanical stress and glucose concentration modulate glucose transport in cultured rat podocytes. Nephrology Dialysis Transplantation, 20(2) , 306-311. -Which statement about glucose transport under physiological conditions is consistent with the kinetic data provided in the passage? A) GLUT2-mediated uptake is a first-order process. B) ATP is required for glucose uptake by both GLUT transporters. C) In the fed state, glucose concentration limits GLUT4-mediated uptake. D) Glucose transport is independent of the concentration gradient.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0e7c_b0ae_a3bf_6191410a37d4_MD0008_00.jpg) Figure 2 Plot of glucose uptake in podocytes with and without mechanical stress.
Adapted from Lewko, B., Bryl, E., Witkowski, J. M., Latawiec, E., Angielski, S., & Stepinski, J. (2005) . Mechanical stress and glucose concentration modulate glucose transport in cultured rat podocytes. Nephrology Dialysis Transplantation, 20(2) , 306-311.
-Which statement about glucose transport under physiological conditions is consistent with the kinetic data provided in the passage?
Figure 2 Plot of glucose uptake in podocytes with and without mechanical stress.
Adapted from Lewko, B., Bryl, E., Witkowski, J. M., Latawiec, E., Angielski, S., & Stepinski, J. (2005) . Mechanical stress and glucose concentration modulate glucose transport in cultured rat podocytes. Nephrology Dialysis Transplantation, 20(2) , 306-311.
-Which statement about glucose transport under physiological conditions is consistent with the kinetic data provided in the passage?
A) GLUT2-mediated uptake is a first-order process.
B) ATP is required for glucose uptake by both GLUT transporters.
C) In the fed state, glucose concentration limits GLUT4-mediated uptake.
D) Glucose transport is independent of the concentration gradient.
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Upon entering muscle cells, glucose is immediately phosphorylated by hexokinase to produce glucose 6-phosphate (G6P) . This phosphorylation event prevents glucose from exiting the cell. G6P may then enter one of three metabolic pathways, depending on the needs of the cell (Figure 1) .
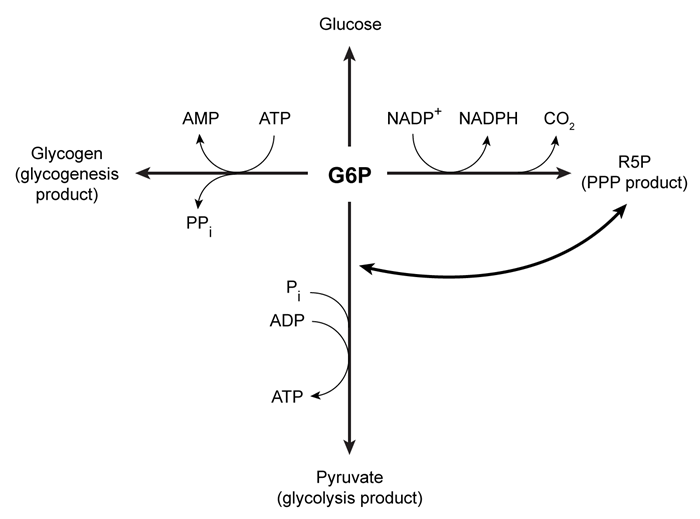 Figure 1 Three potential fates of G6P, determined by the needs of the cellUnder low ATP conditions, G6P enters glycolysis, where it is converted to two pyruvate molecules. The energy released in this process produces a net total of two ATP molecules per glucose molecule consumed. The resulting pyruvate molecules can then enter the mitochondria, where they participate in additional metabolic reactions that produce more ATP.When the cell experiences oxidative stress, G6P enters the pentose phosphate pathway (PPP) and produces NADPH, a reducing agent. During this process, G6P loses one carbon atom in the form of CO2 and is converted to ribose 5-phosphate (R5P) , which is required for nucleotide synthesis. In addition to low NADPH conditions, G6P may also enter the PPP during replication, when nucleotides are needed for DNA synthesis. If nucleotides are not needed but energy is, R5P can be converted back to glycolytic intermediates and reenter glycolysis. Each glucose molecule that is converted to R5P prior to glycolysis yields an average of 1.67 ATP molecules.When all the cell's metabolic requirements are met, additional G6P can enter glycogenesis and produce glycogen, which stores glucose for later use; this process consumes ATP. When signal molecules bind G protein-coupled receptors on muscle cells, glycogenolysis is activated to release individual monomers from glycogen, allowing them to enter the appropriate pathway.
-Which glycolysis intermediate could serve as the sole starting material for the synthesis of R5P in muscle tissue?
Figure 1 Three potential fates of G6P, determined by the needs of the cellUnder low ATP conditions, G6P enters glycolysis, where it is converted to two pyruvate molecules. The energy released in this process produces a net total of two ATP molecules per glucose molecule consumed. The resulting pyruvate molecules can then enter the mitochondria, where they participate in additional metabolic reactions that produce more ATP.When the cell experiences oxidative stress, G6P enters the pentose phosphate pathway (PPP) and produces NADPH, a reducing agent. During this process, G6P loses one carbon atom in the form of CO2 and is converted to ribose 5-phosphate (R5P) , which is required for nucleotide synthesis. In addition to low NADPH conditions, G6P may also enter the PPP during replication, when nucleotides are needed for DNA synthesis. If nucleotides are not needed but energy is, R5P can be converted back to glycolytic intermediates and reenter glycolysis. Each glucose molecule that is converted to R5P prior to glycolysis yields an average of 1.67 ATP molecules.When all the cell's metabolic requirements are met, additional G6P can enter glycogenesis and produce glycogen, which stores glucose for later use; this process consumes ATP. When signal molecules bind G protein-coupled receptors on muscle cells, glycogenolysis is activated to release individual monomers from glycogen, allowing them to enter the appropriate pathway.
-Which glycolysis intermediate could serve as the sole starting material for the synthesis of R5P in muscle tissue?
A) Fructose 6-phosphate
B) Fructose 1,6-bisphosphate
C) Glyceraldehyde 3-phosphate
D) Dihydroxyacetone phosphate
Correct Answer

verified
Correct Answer
verified
Multiple Choice
Passage
Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate.
![Passage Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate. <strong>Figure 1</strong> AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.<strong>Table 1</strong> ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43. -What best describes the action of PH on AMPK activity? A) Long-term agonist B) Immediate antagonist C) Time-dependent antagonist D) Rapid agonist](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0dc2_af71_a3bf_719de41c80b7_MD0008_00.jpg) Figure 1 AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.Table 1 ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo
Figure 1 AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.Table 1 ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo
![Passage Leptin signaling is vital for maintaining adequate body mass and preventing insulin resistance. When energy stores are abundant, adipocytes produce leptin to stimulate fatty acid oxidation and glucose utilization. One result of leptin signaling in skeletal muscle cells is the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK) . AMPK facilitates the breakdown of triglycerides by decreasing the activity of acetyl-coenzyme A carboxylase (ACC) ; this leads to downregulation of fatty acid synthesis and upregulation of β-oxidation.Fatty acid oxidation has been studied by analyzing the effect of leptin on activation of AMPK and ACC in soleus muscles. To identify leptin's mode of action, injections were administered to transgenic mice via the intravenous (1 mg/kg i.v.) and intrahypothalamic (1 ng i.h.p [faster drug delivery]) routes. Phentolamine (PH) , an α-adrenergic receptor ligand that binds to the same receptor as leptin, was administered concurrently in some of the mice. Signaling to ACC from the citric acid cycle was also examined through the addition of citrate. <strong>Figure 1</strong> AMPK activity measured in soleus muscle injected with leptin and PH via i.v. and i.h.p.<strong>Table 1</strong> ACC Activity in Soleus Muscle Exposed to Leptin Delivered via i.v. and i.h.p Injections in vivo Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43. -What best describes the action of PH on AMPK activity? A) Long-term agonist B) Immediate antagonist C) Time-dependent antagonist D) Rapid agonist](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0dc2_af72_a3bf_e9a0f8dccdf8_MD0008_00.jpg) Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43.
-What best describes the action of PH on AMPK activity?
Adapted from Minokoshi Y, Kim YB, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869) :339-43.
-What best describes the action of PH on AMPK activity?
A) Long-term agonist
B) Immediate antagonist
C) Time-dependent antagonist
D) Rapid agonist
Correct Answer

verified
Correct Answer
verified
Showing 21 - 40 of 307